radis.spectrum.spectrum module¶
Summary¶
Spectrum class holder
Contains both the emission and absorption features calculated by a SpectrumFactory, or generated by another spectral code or an experiment and imported in RADIS. Allows use post-processing functions such as applying an instrumental slit, saving to disk, plot all spectral quantities with arbitrary units.
See the The Spectrum object for the list of all post-processing functions, how to save a Spectrum, or how to generate a Spectrum from text.
Examples
Typical use:
from radis calculated_spectrum
s = calculated_spectrum(w, I, conditions={'case':'previously calculated by ##'})
s.plot('radiance_noslit')
s.apply_slit(0.5, shape='triangular')
s.plot('radiance')
Spectrum objects can be modified, stored, resampled, rescaled, or retrieved after
they have been created, with
store(),
rescale_path_length(),
rescale_mole_fraction(),
resample(),
store(),
load_spec()
from radis import load_spec
s = load_spec('co_calculation.spec')
s.rescale_path_length(0.5) # calculate for new path_length
s.rescale_mole_fraction(0.02) # calculate for new mole fraction
s.resample(w_new) # resample on new wavespace
s.store('co_calculation2.spec')
More in The Spectrum object.
- class Spectrum(quantities, units=None, conditions=None, cond_units=None, populations=None, lines=None, wunit=None, name=None, references={}, check_wavespace=True, **kwargs)[source]¶
Bases:
objectThis class holds results calculated with the
SpectrumFactorycalculation, with other radiative codes, or experimental data. It can be used to plot different quantities a posteriori, or manipulate output units (for instance convert a spectral radiance per wavelength units to a spectral radiance per wavenumber).See more information on how to generate, edit or combine Spectrum objects on the Spectrum object guide.
- Parameters:
quantities (dict of tuples
{'quantity':(w, a)}or dict{'wavelength/wavenumber': w, quantity': a}) – where quantities are spectral quantities (absorbance, radiance, etc.) and wavenum is in \(cm^{-1}\) or \(nm\) (seewaveunit) Example:# w, k, I are numpy arrays for wavenumbers, absorption coefficient, and radiance. from radis import Spectrum s = Spectrum({"wavenumber":w, "abscoeff":k, "radiance_noslit":I}, wunit='cm-1', units={"radiance_noslit":"mW/cm2/sr/nm", "abscoeff":"cm-1"})
Or:
s = Spectrum({"abscoeff":(w,k), "radiance_noslit":(w,I)}, wunit="cm-1" units={"radiance_noslit":"mW/cm2/sr/nm", "abscoeff":"cm-1"})
See also:
from_array()andfrom_txt()units (dict) – units for quantities
- Other Parameters:
conditions (dict) – physical conditions and calculation parameters
wunit (
'nm','cm-1','nm_vac'orNone) – wavelength in air ('nm'), wavenumber ('cm-1'), or wavelength in vacuum ('nm_vac'). IfNone,'wavespace'must be defined inconditions. Quantities should be evenly distributed along this space for fast convolution with the slit functioncond_units (dict) – units for conditions
- Other Parameters:
name (str, or None) – Give a name to this Spectrum object (automatically used in plots; useful for multislab configurations). Default
Nonepopulations (dict) – a dictionary of all species, and levels. Should be compatible with other radiative codes such as Specair output. Suggested format: {molecules: {isotopes: {elec state: rovib levels}}} e.g:
{'CO2':{1: 'X': df}} # with df a Pandas Dataframe
lines (pandas Dataframe) – all lines in databank (necessary for using
line_survey()). Warning if you want to play with the lines content: The signification of columns inlinesmay be specific to a database format. Plus, some additional columns may have been added by the calculation (e.g:EiandSfor emission integral and linestrength in SpectrumFactory). Refer to the code to know what they mean (and their units)references (dict) – a dict of
doiof references used to compute this object. Automatically returned with the full bibtex entry bycite()It can also be set a posteriori. Examples = Spectrum() s.references = {"10.1016/j.jqsrt.2010.05.001": "HITEMP-2010 database", "10.1016/j.jqsrt.2018.09.027":["calculation", "post-processing"], # RADIS main paper. Automatically added "10.1016/j.jqsrt.2020.107476":"DIT algorithm"} ) s.cite()
Returns :
Used for DIT algorithm ---------------------- @article{van_den_Bekerom_2021, doi = {10.1016/j.jqsrt.2020.107476}, url = {https://doi.org/10.1016%2Fj.jqsrt.2020.107476}, year = 2021, month = {mar}, publisher = {Elsevier {BV}}, volume = {261}, pages = {107476}, author = {D.C.M. van den Bekerom and E. Pannier}, title = {A discrete integral transform for rapid spectral synthesis}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} } Used for HITEMP-2010 database ----------------------------- @article{Rothman_2010, doi = {10.1016/j.jqsrt.2010.05.001}, url = {https://doi.org/10.1016%2Fj.jqsrt.2010.05.001}, year = 2010, month = {oct}, publisher = {Elsevier {BV}}, volume = {111}, number = {15}, pages = {2139--2150}, author = {L.S. Rothman and I.E. Gordon and R.J. Barber and H. Dothe and R.R. Gamache and A. Goldman and V.I. Perevalov and S.A. Tashkun and J. Tennyson}, title = {{HITEMP}, the high-temperature molecular spectroscopic database}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} } Used for calculation, post-processing ------------------------------------- @article{Pannier_2019, doi = {10.1016/j.jqsrt.2018.09.027}, url = {https://doi.org/10.1016%2Fj.jqsrt.2018.09.027}, year = 2019, month = {jan}, publisher = {Elsevier {BV}}, volume = {222-223}, pages = {12--25}, author = {Erwan Pannier and Christophe O. Laux}, title = {{RADIS}: A nonequilibrium line-by-line radiative code for {CO}2 and {HITRAN}-like database species}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} }
</details>
warnings (boolean) – if
True, test if inputs are valid, e.g, spectra are evenly distributed in wavelength, and raise a warning if not. Note that this take ~ 3.5 ms for a 20k points spectrum, when the rest of the creation process is only ~ 1.8ms (makes it 3 times longer, and can be a problem if hundreds of spectra are created in a row). DefaultTrue
Examples
Manipulate a Spectrum calculated by RADIS:
s = calc_spectrum(2125, 2300, Tgas=2000, databank='CDSD') s.print_conditions() s.plot('absorbance') s.line_survey(overlay='absorbance') s.plot('radiance_noslit', wunits='cm-1', Iunits='W/m2/sr/cm-1') s.apply_slit(5) s.plot('radiance') w, t = s.get('transmittance_noslit') # for use in multi-slabs configs
Any tuple of numpy arrays (w, I) can also be converted into a Spectrum object from the
Spectrumclass directly, or using thecalculated_spectrum()function. All the following methods are equivalent:from radis import Spectrum, calculated_spectrum s1 = calculated_spectrum(w, I, wunit='nm', Iunit='mW/cm2/sr/nm') s2 = Spectrum.from_array(w, I, 'radiance_noslit', wunit='nm', unit='mW/cm2/sr/nm') s3 = Spectrum({'radiance_noslit': (w, I)}, units={'radiance_noslit':'mW/cm2/sr/nm'}, wunit='nm')
See more examples in the [Spectrum] page.
Spectrum objects can be stored, retrieved, rescaled, resampled:
from radis import load_spec s = load_spec('co_calculation.spec') s.rescale_path_length(0.5) # calculate for new path_length s.rescale_mole_fraction(0.02) # calculate for new mole fraction s.resample(w_new) # resample on new wavespace s.store('co_calculation2.spec')
Notes
Implementation:
quantities are stored in the
self._qdictionary. They are better accessed with theget()method that deals with units and wavespaceWavebase:
- c[source]¶
convenience wrapper to
conditions:s.c["calculation_time"] is s.conditions["calculation_time"] >> True
- Type:
dict
- populations[source]¶
Stores molecules, isotopes, electronic states and vibrational or rovibrational populations
- Type:
dict
See also
calculated_spectrum(),transmittance_spectrum(),experimental_spectrum(),from_array(),from_txt(),load_spec()References
[Spectrum]See the Spectrum object page
- apply_slit(slit_function, unit='nm', shape: ['triangular', 'trapezoidal', 'gaussian'] = 'triangular', center_wavespace=None, norm_by='area', mode='valid', plot_slit=False, store=True, slit_dispersion=None, slit_dispersion_threshold=0.01, auto_recenter_crop=True, assert_evenly_spaced=True, verbose=True, inplace=True, *args, **kwargs)[source]¶
Apply an instrumental slit function to all quantities in Spectrum. Slit function can be generated with usual shapes (see
shape=) or imported from an experimental slit function (path to a text file or numpy array of shape n*2). Convoluted spectra are cut on the edge compared to non-convoluted spectra, to remove side effects. Seemode=to change this behaviour.Warning with units: read about
'unit'and'return_unit'parameters.- Parameters:
slit_function (float or str or array) –
- If
float: generate slit function with FWHM of slit function (in nm or cm-1 depending on
unit=). A (top, base) tuple of (float,float) is required when asking for a trapezoidal slit function.- If
.txt: import experimental slit function from .txt file: format must be 2-columns with wavelengths and intensity (doesn’t have to be normalized)
- If
array: format must be 2-columns with wavelengths and intensity (doesn’t have to be normalized) It is recommended to truncate the input slit function to its minimum useful spectral extension (see Notes of
convolve_with_slit()).
- If
unit (
'nm'or'cm-1') – unit of slit_function (FWHM, or imported file)shape (
'triangular','trapezoidal','gaussian', or any ofSLIT_SHAPES) –- which shape to use when generating a slit. Will call,
respectively,
triangular_slit(),trapezoidal_slit(),gaussian_slit(). Default ‘triangular’
center_wavespace (float, or
None) – center of slit when generated (in unit). Not used if slit is imported.norm_by (
'area','max') – normalisation type: -'area'normalizes the slit function to an areaof 1. It conserves energy, and keeps the same units.
'max'normalizes the slit function to a maximum of 1. The convoluted spectrum units change (they are multiplied by the spectrum waveunit, e.g: a radiance non convoluted in mW/cm2/sr/nm on a wavelength (nm). range will yield a convoluted radiance in mW/cm2/sr. Note that the slit is set to 1 in the Spectrum wavespace (i.e: a Spectrum calculated in cm-1 will have a slit set to 1 in cm-1).
Default
'area'mode (
'valid','same') –'same'returns output of same length as initial spectra,but boundary effects are still visible.
'valid'returns output of length len(spectra) - len(slit) + 1, for which lines outside of the calculated range have no impact. Default'valid'.
- Other Parameters:
assert_evenly_spaced (boolean, or
'resample') – for the convolution to be accurate,wshould be evenly spaced. Ifassert_evenly_spaced=True, then we check this is the case, and raise an error if arrays is not evenly spaced. If'resample', then we resamplewandIif needed. Recommended, but it takes some time.auto_recenter_crop (bool) – if
True, recenter slit and crop zeros on the side when importing an experimental slit. DefaultTrue. Seerecenter_slit(),crop_slit()plot_slit (boolean) – if
True, plot slitstore (boolean) – if
True, store slit in the Spectrum object so it can be retrieved withget_slit()and plot withplot_slit(). DefaultTrueslit_dispersion (func of (lambda, in
'nm'), orNone) – spectrometer reciprocal function : dλ/dx(λ) (innm) If notNone, then the slit_dispersion function is used to correct the slit function for the whole range. Can be important if slit function was measured far from the measured spectrum (e.g: a slit function measured at 632.8 nm will look broader at 350 nm because the spectrometer dispersion is higher at 350 nm. Therefore it should be corrected) DefaultNoneWarning
slit dispersion function is assumed to be given in
nmif your spectrum is stored incm-1the wavenumbers are converted to wavelengths before being forwarded to the dispersion functionSee
test_auto_correct_dispersion()for an example of the slit dispersion effect.A Python implementation of the slit dispersion:
>>> def f(lbd): >>> return w/(2*f)*(tan(Φ)+sqrt((2*d/m/(w*1e-9)*cos(Φ))^2-1))
Theoretical / References:
>>> dλ/dx ~ d/mf # at first order >>> dλ/dx = w/(2*f)*(tan(Φ)+sqrt((2*d/m/(w)*cos(Φ))^2-1)) # cf
with:
Φ: spectrometer angle (°)
f: focal length (mm)
m: order of dispersion
d: grooves spacing (mm) = 1/gr with gr in (gr/mm)
See Laux 1999 “Experimental study and modeling of infrared air plasma radiation” for more information
slit_dispersion_warning_threshold (float) – if not
None, check that slit dispersion is about constant (<thresholdchange) on the calculated range. Default 0.01 (1%). Seeoffset_dilate_slit_function()inplace (bool) – if
True, adds convolved arrays directly in the Spectrum. IfFalse, returns a new Spectrum with only the convolved arrays. Note: if you want a new Spectrum with both the convolved and non convolved quantities, uses.copy().apply_slit()
*args, **kwargs – are forwarded to slit generation or import function
verbose (bool) – print stuff
energy_threshold (float) – tolerance fraction when resampling. Default
1e-3(0.1%) If areas before and after resampling differ by more than that an error is raised.
- Returns:
Spectrum – Allows chaining. If
inplace=False, return a new Spectrum with the new spectral arrays only.- Return type:
same Spectrum, with new spectral arrays.
Notes
Units:
the slit function is first converted to the wavespace (wavelength/wavenumber) that the Spectrum is stored in, and applied to the spectral quantities in their native wavespace.
Implementation:
convolve_with_slit()is applied to all quantities inget_vars()that ends with _noslit. Generate a triangular instrumental slit function (or any other shape depending of shape=) with baseslit_function_base(Uses the central wavelength of the spectrum for the slit function generation)We deal with several special cases (which makes the code a little heavy, but the method very versatile):
when slit unit and spectrum unit arent the same
when spectrum is not evenly spaced
Examples
s.apply_slit(1.2, 'nm')
This applies the instrumental function to all available spectral arrays. To manually apply the slit to a particular spectral array, use
take()s.take('transmittance_noslit').apply_slit(1.2, 'nm')
See
convolve_with_slit()for more details on Units and NormalizationThe slit is made considering the “center wavelength” which is the mean wavelength of the full spectrum you are applying it to.
Examples using
radis.Spectrum.apply_slit¶
- argmax(value_only=False)[source]¶
Return wave position of maximum of the Spectrum
Equivalent to the following use of the function
max():s.max(value_only=value_only, return_wmax=True)[1]
See also
- argmin(value_only=False)[source]¶
Return wave position of minimum of the Spectrum
Equivalent to the following use of the function
min():s.min(value_only=value_only, return_wmin=True)[1]
See also
- cite(format='bibentry')[source]¶
Prints bibliographic references used to compute this spectrum, as stored in the
referencesdictionary. Default references known to RADIS are listed inradis.db.references.doi.- Parameters:
format (default
'bibentry'. See more inhabanero.content_negotiation())
Examples
from radis import calc_spectrum s = calc_spectrum( 1900, 2300, # cm-1 molecule="CO", isotope="1,2,3", pressure=1.01325, # bar Tvib=2000, # Trot=300, mole_fraction=0.1, path_length=1, # cm databank="hitran", ) s.cite()
Returns :
Used for algorithm ------------------ @article{van_den_Bekerom_2021, doi = {10.1016/j.jqsrt.2020.107476}, url = {https://doi.org/10.1016%2Fj.jqsrt.2020.107476}, year = 2021, month = {mar}, publisher = {Elsevier {BV}}, volume = {261}, pages = {107476}, author = {D.C.M. van den Bekerom and E. Pannier}, title = {A discrete integral transform for rapid spectral synthesis}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} } Used for calculation, rovibrational energies -------------------------------------------- @article{Pannier_2019, doi = {10.1016/j.jqsrt.2018.09.027}, url = {https://doi.org/10.1016%2Fj.jqsrt.2018.09.027}, year = 2019, month = {jan}, publisher = {Elsevier {BV}}, volume = {222-223}, pages = {12--25}, author = {Erwan Pannier and Christophe O. Laux}, title = {{RADIS}: A nonequilibrium line-by-line radiative code for {CO}2 and {HITRAN}-like database species}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} } Used for data retrieval ----------------------- @article{Ginsburg_2019, doi = {10.3847/1538-3881/aafc33}, url = {https://doi.org/10.3847%2F1538-3881%2Faafc33}, year = 2019, month = {feb}, publisher = {American Astronomical Society}, volume = {157}, number = {3}, pages = {98}, author = {Adam Ginsburg and Brigitta M. Sip{\H{o}}cz and C. E. Brasseur and Philip S. Cowperthwaite and Matthew W. Craig and Christoph Deil and James Guillochon and Giannina Guzman and Simon Liedtke and Pey Lian Lim and Kelly E. Lockhart and Michael Mommert and Brett M. Morris and Henrik Norman and Madhura Parikh and Magnus V. Persson and Thomas P. Robitaille and Juan-Carlos Segovia and Leo P. Singer and Erik J. Tollerud and Miguel de Val-Borro and Ivan Valtchanov and Julien Woillez and}, title = {astroquery: An Astronomical Web-querying Package in Python}, journal = {The Astronomical Journal} } Used for line database ---------------------- @article{Gordon_2017, doi = {10.1016/j.jqsrt.2017.06.038}, url = {https://doi.org/10.1016%2Fj.jqsrt.2017.06.038}, year = 2017, month = {dec}, publisher = {Elsevier {BV}}, volume = {203}, pages = {3--69}, author = {I.E. Gordon and L.S. Rothman and C. Hill and R.V. Kochanov and Y. Tan and P.F. Bernath and M. Birk and V. Boudon and A. Campargue and K.V. Chance and B.J. Drouin and J.-M. Flaud and R.R. Gamache and J.T. Hodges and D. Jacquemart and V.I. Perevalov and A. Perrin and K.P. Shine and M.-A.H. Smith and J. Tennyson and G.C. Toon and H. Tran and V.G. Tyuterev and A. Barbe and A.G. Cs{\'{a}}sz{\'{a}}r and V.M. Devi and T. Furtenbacher and J.J. Harrison and J.-M. Hartmann and A. Jolly and T.J. Johnson and T. Karman and I. Kleiner and A.A. Kyuberis and J. Loos and O.M. Lyulin and S.T. Massie and S.N. Mikhailenko and N. Moazzen-Ahmadi and H.S.P. Müller and O.V. Naumenko and A.V. Nikitin and O.L. Polyansky and M. Rey and M. Rotger and S.W. Sharpe and K. Sung and E. Starikova and S.A. Tashkun and J. Vander Auwera and G. Wagner and J. Wilzewski and P. Wcis{\l}o and S. Yu and E.J. Zak}, title = {The {HITRAN}2016 molecular spectroscopic database}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} } Used for partition function --------------------------- @article{Gamache_2021, doi = {10.1016/j.jqsrt.2021.107713}, url = {https://doi.org/10.1016%2Fj.jqsrt.2021.107713}, year = 2021, month = {sep}, publisher = {Elsevier {BV}}, volume = {271}, pages = {107713}, author = {Robert R. Gamache and Bastien Vispoel and Michaël Rey and Andrei Nikitin and Vladimir Tyuterev and Oleg Egorov and Iouli E. Gordon and Vincent Boudon}, title = {Total internal partition sums for the {HITRAN}2020 database}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} } @article{Kochanov_2016, doi = {10.1016/j.jqsrt.2016.03.005}, url = {https://doi.org/10.1016%2Fj.jqsrt.2016.03.005}, year = 2016, month = {jul}, publisher = {Elsevier {BV}}, volume = {177}, pages = {15--30}, author = {R.V. Kochanov and I.E. Gordon and L.S. Rothman and P. Wcis{\l}o and C. Hill and J.S. Wilzewski}, title = {{HITRAN} Application Programming Interface ({HAPI}): A comprehensive approach to working with spectroscopic data}, journal = {Journal of Quantitative Spectroscopy and Radiative Transfer} } Used for spectroscopic constants -------------------------------- @article{Guelachvili_1983, doi = {10.1016/0022-2852(83)90203-5}, url = {https://doi.org/10.1016%2F0022-2852%2883%2990203-5}, year = 1983, month = {mar}, publisher = {Elsevier {BV}}, volume = {98}, number = {1}, pages = {64--79}, author = {G. Guelachvili and D. de Villeneuve and R. Farrenq and W. Urban and J. Verges}, title = {Dunham coefficients for seven isotopic species of {CO}}, journal = {Journal of Molecular Spectroscopy} }
Other Examples¶
See also
- compare_with(other, spectra_only=False, plot=True, wunit='default', verbose=True, rtol=1e-05, ignore_nan=False, ignore_outliers=False, normalize=False, **kwargs)[source]¶
Compare Spectrum with another Spectrum object.
- Parameters:
other (type Spectrum) – another Spectrum to compare with
spectra_only (boolean, or str) – if
True, only compares spectral quantities (in the same waveunit) and not lines or conditions. If str, compare a particular quantity name. If False, compare everything (including lines and conditions and populations). DefaultFalseplot (boolean) – if
True, use plot_diff to plot all quantities for the 2 spectra and the difference between them. DefaultTrue.wunit (
"nm","cm-1","default") – in which wavespace to compare (and plot). If"default", natural wavespace of first Spectrum is taken.rtol (float) – relative difference to use for spectral quantities comparison
ignore_nan (boolean) – if
True, nans are ignored when comparing spectral quantitiesignore_outliers (boolean, or float) –
if not False, outliers are discarded. i.e, output is determined by:
out = (~np.isclose(I, Ie, rtol=rtol, atol=0)).sum()/len(I) < ignore_outliers
normalize (bool) – Normalize the spectra to be plotted
- Other Parameters:
kwargs (dict) – arguments are forwarded to
plot_diff()- Returns:
equals – return True if spectra are equal (respective to tolerance defined by rtol and other input conditions)
- Return type:
boolean
Examples
Compare two Spectrum objects, or specifically the transmittance:
s1.compare_with(s2) s1.compare_with(s2, 'transmittance')
Note that you can also simply use
s1 == s2, that usescompare_with()internally:s1 == s2 # will return True or False
See also
- conditions[source]¶
computation conditions, or experimental parameters, or any metadata you need to store with the Spectrum object.
- Type:
dict
- copy(copy_lines=True, quantity='all', copy_arrays=True)[source]¶
Returns a copy of this Spectrum object (performs a smart deepcopy)
- Parameters:
copy_lines (bool) – default
Truequantity (‘all’, or one of ‘radiance_noslit’, ‘absorbance’, etc.) – if not ‘all’, copy only one quantity. Default
'all'copy_arrays (bool) – if
False, returned array’s quantity is a pointer to the original Spectrum. Faster, but warning, changing them will then change the original Spectrum. DefaultTrue
Examples
- crop(wmin=None, wmax=None, wunit='default', inplace=True)[source]¶
Crop spectrum to
wmin-wmaxrange inwunit(inplace)- Parameters:
wmin, wmax (float, or None) – boundaries of spectral range (in
wunit)wunit (
'nm','cm-1','nm_vac') – which waveunit to use forwmin, wmax. Ifdefault: use the default Spectrum wavespace defined withget_waveunit().
- Other Parameters:
inplace (bool) – if
True, modifies the Spectrum object directly. Else, returns a copy. DefaultTrue.- Returns:
s – Cropped Spectrum. If
inplace=True, Spectrum has been updated directly anyway. Allows chaining- Return type:
Examples
Crop to experimental Spectrum, and compare:
from radis import calc_spectrum, load_spec, plot_diff s = calc_spectrum(...) s_exp = load_spec('typical_result.spec') s.crop(s_exp.get_wavelength.min(), s_exp.get_wavelength.max(), 'nm') plot_diff(s_exp, s)
See also
- fit_model(model, plot=False, confidence=0.9545, verbose=False, debug=False)[source]¶
Fit a lineshape model to the spectrum.
The model can be a simple lineshape model among
Gaussian1D,Lorentz1D,Voigt1D,or a combination of multiple models.
- Parameters:
model (astropy.modeling.Model or a list of models) – model to fit to the spectrum. If a list is given, a sum of models is fitted
plot (bool) – if True, plot the difference between the model and the spectrum
- Other Parameters:
confidence (0.6827, 0.9545, or 0.9973) – confidence interval to use.
- Returns:
g_fit – the fitted model
y_err – uncertainty on the fitted parameters calculated as the square root of the diagonal of the covariance matrix
Examples
from astropy.modeling import models s = radis.test_spectrum().crop(2201.7, 2205.1) g_fit, y_err = s.fit_model(models.Lorentz1D(), plot=True)
Example with 6 Voigt lines:
from astropy.modeling import models s = radis.test_spectrum().crop(2201.7, 2225.1) g_fit_list, y_err = s.fit_model([models.Voigt1D() for _ in range(6)], plot=True)
Other Examples¶
- classmethod from_array(w, I, quantity, wunit=None, Iunit=None, waveunit=None, unit=None, *args, **kwargs)[source]¶
Construct Spectrum from 2 arrays.
- Parameters:
w, I (array) – waverange and vector
quantity (str) – spectral quantity name
wunit (
'nm','cm-1','nm_vac') – unit of waverange: wavelength in air ('nm'or'nm_air'), wavenumber ('cm-1'), or wavelength in vacuum ('nm_vac'). IfNone, thenwmust be a dimensioned array.Iunit (str) – spectral quantity unit (arbitrary). Ex:
'mW/cm2/sr/nm'for radiance_noslit IfNone, thenImust be a dimensioned array.*args, **kwargs – see
Spectrumdoc
- Other Parameters:
conditions (dict) – physical conditions and calculation parameters
cond_units (dict) – units for conditions
populations (dict) – a dictionary of all species, and levels. Should be compatible with other radiative codes such as Specair output. Suggested format: {molecules: {isotopes: {elec state: rovib levels}}} e.g:
{'CO2':{1: 'X': df}} # with df a Pandas Dataframe
lines (pandas Dataframe) – all lines in databank (necessary for using
line_survey()). Warning if you want to play with the lines content: The signification of columns inlinesmay be specific to a database format. Plus, some additional columns may have been added by the calculation (e.g:EiandSfor emission integral and linestrength in SpectrumFactory). Refer to the code to know what they mean (and their units)
- Returns:
creates a
Spectrumobject- Return type:
Examples
Create a spectrum:
from radis import Spectrum s = Spectrum.from_array(w, I, 'radiance_noslit', wunit='nm', unit='mW/cm2/sr/nm')
Dimensioned arrays can also be used directly
import astropy.units as u w = np.linspace(200, 300) * u.nm I = np.random.rand(len(w)) * u.mW/u.cm**2/u.sr/u.nm s = Spectrum.from_array(w, I, 'radiance_noslit')
To create a spectrum with absorption and emission components (e.g:
radiance_noslitandtransmittance_noslit, oremisscoeffandabscoeff) call theSpectrumclass directly. Ex:from radis import Spectrum s = Spectrum({'abscoeff': (w, A), 'emisscoeff': (w, E)}, units={'abscoeff': 'cm-1', 'emisscoeff':'W/cm2/sr/nm'}, wunit='nm')
- classmethod from_hdf5(file, wmin=None, wmax=None, wunit=None, columns=None, engine='pytables')[source]¶
Generates a Spectrum from an HDF5 file. Uses
hdf2spec()- Other Parameters:
wmin, wmax (float) – range of wmin, wmax to load , using
wunit(if None, load everything)columns (list of str) – spectral arrays to load (if None, load everything)
Examples
Spectrum.from_hdf5("rad_hdf.h5", wmin=2100, wmax=2200, columns=['abscoeff', 'emisscoeff'])
See also
- classmethod from_mat(file, quantity, wunit, unit, data_key=None, w_name='nu', I_name=None, index=None, *args, **kwargs)[source]¶
Construct Spectrum from Matlab
.matfile.- Parameters:
file (str) – file name
quantity (str) – spectral quantity name
wunit (
'nm','cm-1','nm_vac') – unit of waverange: wavelength in air ('nm'), wavenumber ('cm-1'), or wavelength in vacuum ('nm_vac').unit (str) – spectral quantity unit
index (int) – index within Matlab
.matarray.data_key (str) – data key to use within Matlab
.matarray. IfNone, guess.w_name, I_name (str) – key to use to parse the
data[data_key]array and return waverange and quantity.*args, **kwargs – the following inputs are forwarded to
loadmat():'simplify_cells','skiprows'The rest if forwarded to Spectrum and will be registered as a Spectrum condition. seeSpectrumdoc
- Returns:
s – creates a
Spectrumobject- Return type:
Examples
Notes
Internally, the scipy
loadmat()function is used and corresponds todata = scipy.io.loadmat(file, **kwloadmat) data = data[data_key] w, I = data[w_name], data[I_name]
Special keywords can be given to
kwloadmat. See docs ofkwargs.See also
- classmethod from_spec(file)[source]¶
Generates a Spectrum from a .spec [json] file. Uses
load_spec()
- classmethod from_specutils(spectrum, var='radiance')[source]¶
Convert a
specutilsSpectrum1Dto aradisSpectrumobject.- Parameters:
spectrum (a
specutilsspecutils.spectra.spectrum1d.Spectrum1D)var (str) – spectral array, default
"radiance"
Examples
Taken from the Specutils website (https://specutils.readthedocs.io/en/stable/#getting-started-with-specutils)
from astropy.io import fits from astropy import units as u from specutils import Spectrum1D f = fits.open('https://data.sdss.org/sas/dr16/sdss/spectro/redux/26/spectra/1323/spec-1323-52797-0012.fits') # The spectrum is in the second HDU of this file. specdata = f[1].data lamb = 10**specdata['loglam'] * u.AA flux = specdata['flux'] * 10**-17 * u.Unit('erg cm-2 s-1 AA-1') spec = Spectrum1D(spectral_axis=lamb, flux=flux) from radis import Spectrum s = Spectrum.from_specutils(spec) s.plot(wunit='nm')
See also
- classmethod from_txt(file, quantity, wunit, unit, waveunit=None, *args, **kwargs)[source]¶
Construct Spectrum from txt file.
- Parameters:
file (str) – file name
quantity (str) – spectral quantity name
wunit (
'nm','cm-1','nm_vac') – unit of waverange: wavelength in air ('nm'), wavenumber ('cm-1'), or wavelength in vacuum ('nm_vac').unit (str) – spectral quantity unit
*args, **kwargs – the following inputs are forwarded to loadtxt:
'delimiter','skiprows'The rest if forwarded to Spectrum. seeSpectrumdoc
- Other Parameters:
delimiter (
',', etc.) – seenumpy.loadtxt()skiprows (int) – see
numpy.loadtxt()argsort (bool) – sorts the arrays in
fileby wavespace. Convenient way to load a file where points have been manually added at the end. DefaultFalse.*Optional Spectrum parameters*
conditions (dict) – physical conditions and calculation parameters
cond_units (dict) – units for conditions
populations (dict) – a dictionary of all species, and levels. Should be compatible with other radiative codes such as Specair output. Suggested format: {molecules: {isotopes: {elec state: rovib levels}}} e.g:
{'CO2':{1: 'X': df}} # with df a Pandas Dataframe
lines (pandas Dataframe) – all lines in databank (necessary for using
line_survey()). Warning if you want to play with the lines content: The signification of columns inlinesmay be specific to a database format. Plus, some additional columns may have been added by the calculation (e.g:EiandSfor emission integral and linestrength in SpectrumFactory). Refer to the code to know what they mean (and their units)
- Returns:
s – creates a
Spectrumobject- Return type:
Examples
Generate an experimental spectrum from txt. In that example the
delimiterkey is forwarded toloadtxt():from radis import Spectrum s = Spectrum.from_txt('spectrum.csv', 'radiance', wunit='nm', unit='W/cm2/sr/nm', delimiter=',')
To create a spectrum with absorption and emission components (e.g:
radiance_noslitandtransmittance_noslit, oremisscoeffandabscoeff) call theSpectrumclass directly. Ex:from radis import Spectrum s = Spectrum({'abscoeff': (w, A), 'emisscoeff': (w, E)}, units={'abscoeff': 'cm-1', 'emisscoeff':'W/cm2/sr/nm'}, wunit='nm')
Notes
Internally, the numpy
loadtxt()function is used and transposed:w, I = np.loadtxt(file).T
You can use
'delimiter'and ‘skiprows'as arguments.
- generate_perf_profile()[source]¶
Generate a visual/interactive performance profile diagram using
tunaNote
requires a
profilerkey with in Spectrum.conditionsWarning
deprecated in favor of
print_perf_profile()Examples
s = calc_spectrum(...) s.generate_perf_profile()
See typical output in https://github.com/radis/radis/pull/325

Note
You can also profile with
tunadirectly:python -m cProfile -o program.prof your_radis_script.py tuna your_radis_script.py
See also
- get(var: ['abscoeff', 'absorbance', 'emissivity_noslit', 'transmittance_noslit', 'radiance_noslit'], wunit='default', Iunit='default', copy=True, trim_nan=False, return_units=False)[source]¶
Retrieve a spectral quantity from a Spectrum object. You can select wavespace unit, intensity unit, or propagation medium.
- Parameters:
var (variable (
'absorbance','transmittance','xsection'etc.)) – Should be a defined quantity amongCONVOLUTED_QUANTITIESorNON_CONVOLUTED_QUANTITIES. To get the full list of spectral arrays defined in this Spectrum object use theget_vars()method.wunit (
'nm','cm','nm_vac'.) – wavespace unit: wavelength in air ('nm'), wavenumber ('cm-1'), or wavelength in vacuum ('nm_vac'). if"default", default unit for waveunit is used. Seeget_waveunit().Iunit (unit for variable
var) – if"default", default unit for quantityvaris used. See theunitsattribute. Forvar="radiance", one can use per wavelength (~ ‘W/m2/sr/nm’) or per wavenumber (~ ‘W/m2/sr/cm-1’) unitsNote
if using
default, the returned unit may be different from Spectrum.units[var]. e.g, for a Spectrum with radiance stored in ‘mW/cm2/sr/nm’, getting it withwunit='cm-1', Iunit='default'will return Iunit in ‘mW/cm2/sr/cm-1’ for consistency. Usereturn_unitsto be sure about which units are used.
- Other Parameters:
copy (bool) – if
True, returns a copy of the stored quantity (modifying it wont change the Spectrum object). DefaultTrue.trim_nan (bool) – if
True, removesnanon the sides of the spectral array (and corresponding wavespace). DefaultFalse.return_units (bool) – if
True, return dimensioned Astropy arrays. Is usingreturn_units = 'as_str', returnwunitandIunitas extra variables, i.e,w, I, wunit, Iunit = s.get(..., return_units='as_str')DefaultFalse
- Returns:
w, I – wavespace, quantity (ex: wavelength, radiance_noslit). For numpy users, note that these are copies (values) of the Spectrum quantity and not a view (reference): if you modify them the Spectrum is not changed
- Return type:
array-like
Examples
Get transmittance in cm-1:
w, I = s.get('transmittance_noslit', wunit='cm-1')
Get radiance (in wavelength in air):
_, R = s.get('radiance_noslit', wunit='nm', Iunit='W/cm2/sr/nm')
Use with
return_unitsto get dimensioned Astropy Quantitiesw, R = s.get('radiance_noslit', return_units=True)
See also
- get_baseline(algorithm='als', **kwargs)[source]¶
Calculate and returns a baseline
- Parameters:
s (Spectrum) – Spectrum which needs a baseline
var (str) – on which spectral quantity to read the baseline. Default
'radiance'. SeeSPECTRAL_QUANTITIESalgorithm (‘als’, ‘polynomial’) – Asymmetric least square or Polynomial fit
**kwargs (dict) – additional parameters to send to the algorithm. By default, for ‘polynomial’:
for ‘als’:
- **kwargs = {“asymmetry_param”: 0.05,
“smoothness_param”: 1e6}
- Returns:
baseline – Spectrum object where intensity is the baseline of s
- Return type:
Examples
See also
- get_integral(var, wunit='default', Iunit='default', **kwargs)[source]¶
Returns integral of variable ‘var’ over waverange.
- Parameters:
var (str) – spectral quantity to integrate
wunit (str) – over which waverange to integrated. If
default, use the default Spectrum wavespace defined withget_waveunit().Iunit (str) – default
'default'Warning
this is the unit of the quantity, not the unit of the integral. Don’t forget to multiply by
wunit
- Other Parameters:
- Returns:
integral – integral in [Iunit]*[wunit]
- Return type:
float
See also
- get_name()[source]¶
Return Spectrum name.
If not defined, returns either the
filename if Spectrum was loaded from a file, or the'spectrum{id}'with the Pythonidobject
- get_populations(molecule=None, isotope=None, electronic_state=None, show_warning=True)[source]¶
Return populations that are featured in the spectrum, either as upper or lower levels.
- Parameters:
molecule (str, or None) – if None, only one molecule must be defined. Else, an error is raised
isotope (int, or None) – isotope number. if None, only one isotope must be defined. Else, an error is raised
electronic_state (str) – if None, only one electronic state must be defined. Else, an error is raised
show_warning (bool) – if False, turns off warning about meaning of populations, see Notes and discussion on https://github.com/radis/radis/issues/508.
- Returns:
pandas dataframe of levels, where levels are the index,
and ‘Evib’ and ‘nvib’ are featured
Notes
Structure:
{molecule: {isotope: {electronic_state: {'vib': pandas Dataframe, # (copy of) vib levels 'rovib': pandas Dataframe, # (copy of) rovib levels 'Ia': float # isotopic abundance }}}}
(If Spectrum generated with RADIS, structure should match that of SpectrumFactory.get_populations())
Examples
An example on how different are populations used for partition function and spectrum calculations
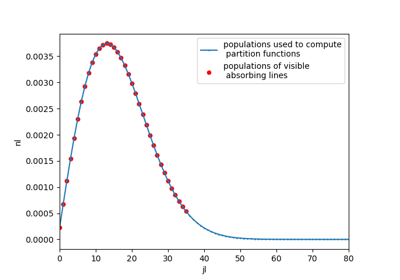
#%% CO2 example # For instance, plot populations of a given vibrational level, v1,v2,v3=(0,1,0) import radis s = radis.test_spectrum(molecule="CO2", Tvib=3000, Trot=1000, export_lines=True, export_populations="rovib", isotope=1) pops = s.get_populations("CO2")["rovib"] import matplotlib.pyplot as plt pops.query("v1==0 & v2==1 & v3==0").plot("j", "n", label="pops. used to compute partition functions") s.lines.query("v1l==0 & v2l==1 & v3l==0").plot("jl", "nl", ax=plt.gca(), kind="scatter", color="r", label="pops. of visible absorbing lines") plt.legend() plt.xlim((0,70))
See also
- get_power(unit='mW/cm2/sr')[source]¶
Returns integrated radiance (no slit) power density.
- Parameters:
Iunit (str) – power unit.
- Returns:
P – radiated power in
unit- Return type:
float
Examples
s.get_power('W/cm2/sr')
See also
- get_quantities(which=None)[source]¶
Returns all spectral quantities stored in this object (convoluted or non convoluted). Wrapper to
get_vars()
- get_radiance(Iunit='mW/cm2/sr/nm', copy=True)[source]¶
Return radiance in whatever unit, and can even convert from ~1/nm to ~1/cm-1 (and the other way round)
- Other Parameters:
copy (boolean) – if
True, returns a copy of the stored waverange (modifying it wont change the Spectrum object). DefaultTrue.
See also
- get_radiance_noslit(Iunit='mW/cm2/sr/nm', copy=True)[source]¶
Return radiance (non convoluted) in whatever unit, and can even convert from ~1/nm to ~1/cm-1 (and the other way round)
- Other Parameters:
copy (boolean) – if
True, returns a copy of the stored waverange (modifying it wont change the Spectrum object). DefaultTrue.
See also
- get_rovib_levels(molecule=None, isotope=None, electronic_state=None, first=None)[source]¶
Return rovibrational levels calculated in the spectrum (energies, populations)
- Parameters:
molecule (str, or None) – if None, only one molecule must be defined. Else, an error is raised
isotope (int, or None) – isotope number. if None, only one isotope must be defined. Else, an error is raised
electronic_state (str) – if None, only one electronic state must be defined. Else, an error is raised
first (int, or ‘all’ or None) – only show the first N levels. If None or ‘all’, all levels are shown
- Returns:
out – pandas dataframe of levels, where levels are the index, and ‘Evib’ and ‘nvib’ are featured
- Return type:
pandas DataFrame
- get_slit(wunit='same')[source]¶
Get slit function that was applied to the Spectrum.
- Returns:
wslit, Islit – slit function with wslit in Spectrum
waveunit. Seeget_waveunit()- Return type:
array
- get_vars(which=None)[source]¶
Returns all spectral quantities stored in this object (convoluted or non convoluted)
- get_vib_levels(molecule=None, isotope=None, electronic_state=None, first=None)[source]¶
Return vibrational levels in the spectrum (energies, populations)
- Parameters:
molecule (str, or None) – if None, only one molecule must be defined. Else, an error is raised
isotope (int, or None) – isotope number. if None, only one isotope must be defined. Else, an error is raised
electronic_state (str) – if None, only one electronic state must be defined. Else, an error is raised
first (int, or ‘all’ or None) – only show the first N levels. If None or ‘all’, all levels are shown
- Returns:
out – pandas dataframe of levels, where levels are the index, and ‘Evib’ and ‘nvib’ are featured
- Return type:
pandas DataFrame
- get_wavelength(medium='air', which=None, copy=True)[source]¶
Return wavelength in defined medium.
- Parameters:
medium (
'air','vacuum') – returns wavelength as seen in air, or vacuum. Default'air'. Seevacuum2air(),air2vacuum()- Other Parameters:
copy (boolean) – if
True, returns a copy of the stored waverange (modifying it wont change the Spectrum object). DefaultTrue.- Returns:
w – (a copy of) spectrum wavelength for convoluted or non convoluted quantities
- Return type:
array_like
See also
- get_wavenumber(which=None, copy=True)[source]¶
Return wavenumber (if the same for all quantities)
- Other Parameters:
copy (boolean) – if
True, returns a copy of the stored waverange (modifying it wont change the Spectrum object). DefaultTrue.- Returns:
w – (a copy of) spectrum wavenumber for convoluted or non convoluted quantities
- Return type:
array_like
- get_waveunit()[source]¶
Returns whether this spectrum is defined in wavelength (nm) or wavenumber (cm-1)
- has_nan(ignore_wavespace=True) bool[source]¶
- Parameters:
s (Spectrum) – radis Spectrum.
- Returns:
b – returns whether Spectrum has
nan- Return type:
bool
- is_at_equilibrium(check='warn', verbose=False)[source]¶
Returns whether this spectrum is at (thermal) equilibrium. Reads the
thermal_equilibriumkey in Spectrum conditions. It does not imply chemical equilibrium (mole fractions are still arbitrary)If they are defined, also check that the following assertions are True:
Tvib = Trot = Tgas self_absorption = True overpopulation = None
If they are not, still trust the value in Spectrum conditions, but raise a warning.
- Other Parameters:
check (
'warn','error','ignore') – what to do if Spectrum conditions dont match the given equilibrium state: raise a warning, raise an error, or just ignore and dont even check. Default'warn'.verbose (bool) – if
True, print why is the spectrum is not at equilibrium, if applicable.
- is_optically_thin()[source]¶
Returns whether the spectrum is optically thin, based on the value on the self_absorption key in conditions.
If not given, raises an error
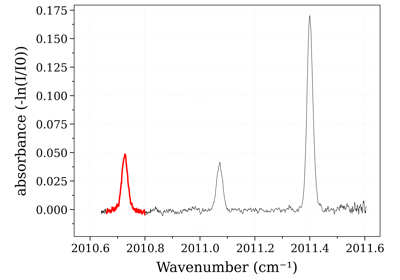
- line_survey(overlay=None, wunit='cm-1', Iunit='hitran', medium='air', cutoff=None, plot='S', lineinfo=['int', 'A', 'El'], barwidth='hwhm_voigt', yscale='log', writefile=None, *args, **kwargs)[source]¶
- Plot Line Survey (all linestrengths used for calculation) Output in
Plotly (html)
- Parameters:
overlay (tuple (w, I, [name], [units]), or list or tuples) – plot (w, I) on a secondary axis. Useful to compare linestrength with calculated / measured data:
LineSurvey(overlay='abscoeff')
wunit (
'nm','cm-1') – wavelength / wavenumber unitsIunit (
'hitran','splot') – Linestrength output units:'hitran': (cm-1/(molecule/cm-2))'splot': (cm-1/atm) (Spectraplot units [2])
Note: if not None, cutoff criteria is applied in this unit. Not used if plot is not ‘S’
medium (
'air','vacuum') – show wavelength either in air or vacuum. Default'air'plot (str) – what to plot. Default
'S'(scaled line intensity). But it can be any key in the lines, such as population ('nu'), or Einstein coefficient ('Aul')lineinfo (list, or
'all') – extra line information to plot. Should be a column name in the databank (s.lines). For instance:'int','selbrd', etc… Default ['int']
- Other Parameters:
- writefile (str) – if not
None, a valid filename to save the plot under .html format. If
None, use thefigobject returned to show the plot.yscale (
'log','linear') – Default'log'barwidth (float or str) –
if float, width of bars, in
wunit, as a fraction of full-range; i.e.barwidth=0.01
makes bars span 1% of the full range. if
str, uses the column as width. Examplebarwidth = 'hwhm_voigt'
- returns:
fig (a Plotly figure.) – If using a Jupyter Notebook, the plot will appear. Else, use
writefileto export to an html file.Plot in Plotly. See Output in [1]_
- writefile (str) – if not
An example using the
SpectrumFactoryto generate a spectrum:from radis import SpectrumFactory sf = SpectrumFactory( wavenum_min=2380, wavenum_max=2400, mole_fraction=400e-6, path_length=100, # cm isotope=[1], export_lines=True, # required for LineSurvey! db_use_cached=True) sf.load_databank('HITRAN-CO2-TEST') s = sf.eq_spectrum(Tgas=1500) s.apply_slit(0.5) s.line_survey(overlay='radiance_noslit', barwidth=0.01, lineinfo="all") # or barwidth='hwhm_voigt'
See the output in Examples
- lines[source]¶
informations on emitting or absorbing lines that contribute to the spectrum.
See also
line_survey()
- max(value_only=False, return_wmax=False)[source]¶
Maximum of the Spectrum, if only one spectral quantity is available:
s.max()
Returns a dimensioned quantity by default, with the default unit of the spectrum. If
value_only=False, a float is returned, without dimensions.If there are multiple arrays in your Spectrum, use
take(), e.g.s.take('radiance').max()
- Parameters:
value_only (bool)
return_position (bool) – if True, returns wavelength or wavenumber of maximum, in the Spectrum unit.
Examples
Normalize a spectrum (we could also have used
normalize()directly)s /= s.max()
Below is a slightly more advanced use case, convenient to process non-calibrated experimental data.
Imagine we want to remove a baseline that we have identified to originate from a particular species, modelled by spectrum
s, but whose intensity (and units) are not the same as that of the experimental spectrum.We substract exactly the intensity that correspond to the intensity on a given range
w1, w2of the experimental spectrums_exp, using a combination oftake(),normalize(),crop()andmax():s_exp -= s.take('radiance').normalize() * s_exp.crop((w1, w2)), inplace=False).max()
Other Examples¶
Get max position
wmaxs = radis.test_spectrum() max, wmax = s.max(return_wmax=True)
Note that the later can also be achieved with
argmax()wmax = s.argmax()
See also
- min(value_only=True, return_wmin=False)[source]¶
Minimum of the Spectrum, if only one spectral quantity is available
s.min()
Returns a dimensioned quantity by default, with the default unit of the spectrum. If
value_only=False, a float is returned, without dimensions.If there are multiple arrays in your Spectrum, use
take(), e.g.s.take('radiance').min()
Other Examples¶
Get min position
wmins = radis.test_spectrum() min, wmin = s.min(return_wmin=True)
Note that the later can also be achieved with
argmin()wmax = s.argmin()
See also
- normalize(normalize_how='max', wrange=(), wunit=None, inplace=False, force=False, verbose=False, return_norm=False)[source]¶
Normalise the Spectrum, if only one spectral quantity is available.
- Parameters:
normalize_how (
'max','area','mean') – how to normalize.'max'is the default but may not be suited for very noisy experimental spectra.'area'will normalize the integral to 1.'mean'will normalize by the mean amplitude valuewrange (tuple of floats, float) – normalize on this range if a tuple, else normalise at that point. If an empty tuple, use the entire range.
wunit (
"nm","cm-1","nm_vac") – unit of the normalisation range above. IfNone, use the spectrum default waveunit.inplace (bool) – if
True, changes the Spectrum.
- Other Parameters:
force (boolean) – By default, normalizing some parameters such as transmittance is forbidden because considered non-physical. Use force=True if you really want to.
Examples
s.normalize("max", (4200, 4800), inplace=True).plot()
If there are multiple arrays in your Spectrum, use
take(), e.g.s.take('radiance').normalize(wrange=(4200, 4800)).plot()
- offset(offset: Spectrum, unit: float, inplace: str = True) Spectrum[source]¶
Offset the spectrum by a wavelength or wavenumber (inplace)
- Parameters:
offset (float) – Constant to add to all quantities in the Spectrum.
unit (‘nm’ or ‘cm-1’) – unit for
offset
- Other Parameters:
inplace (bool) – if
True, modifies the Spectrum object directly. Else, returns a copy. DefaultTrue.- Returns:
s – Offset Spectrum. If
inplace=True, Spectrum has been updated directly anyway. Allows chaining.- Return type:
Examples
s.offset(5, 'nm')
- plot(var: ['abscoeff', 'absorbance', 'emissivity_noslit', 'transmittance_noslit', 'radiance_noslit'] = None, wunit='default', Iunit='default', show_points=False, nfig=None, yscale='linear', show_medium='vacuum_only', normalize=False, force=False, plot_by_parts=False, show=True, show_ruler=False, **kwargs)[source]¶
Plot a
Spectrumobject.Note
default plotting library and templates can be edited in
radis.config[“plot”]- Parameters:
var (variable (
absorbance,transmittance,transmittance_noslit,xsection, etc.)) – For full list seeget_vars(). IfNone, plot the first thing in the Spectrum. DefaultNone.wunit (
'default','nm','cm-1','nm_vac',) – wavelength air, wavenumber, or wavelength vacuum. If'default', Spectrumget_waveunit()is used.Iunit (unit for variable) – if
default, default unit for quantityvaris used. for radiance, one can use per wavelength (~W/m2/sr/nm) or per wavenumber (~W/m2/sr/cm-1) units
- Other Parameters:
show_points (boolean) – show calculated points. Default
True.nfig (int, None, or ‘same’) – plot on a particular figure. ‘same’ plots on current figure. For instance:
s1.plot() s2.plot(nfig='same')
show_medium (bool,
'vacuum_only') – ifTrueandwunitare wavelengths, plot the propagation medium in the xaxis label ([air]or[vacuum]). If'vacuum_only', plot only ifwunit=='nm_vac'. Default'vacuum_only'(prevents from inadvertently plotting spectra with different propagation medium on the same graph).yscale (‘linear’, ‘log’) – plot yscale
normalize (boolean, or tuple.) – option to normalize quantity to 1 (ex: for radiance). Default
Falseplot_by_parts (bool) – if
True, look for discontinuities in the wavespace and plot the different parts without connecting lines. Useful for experimental spectra produced by overlapping step-and-glue. Additional parameters read fromkwargs:split_thresholdandcutwings. See more insplit_and_plot_by_parts().force (bool) – plotting on an existing figure is forbidden if labels are not the same. Use
force=Trueto ignore that.show (bool) – show figure. Default
False. Will still show the figure in interactive mode, e.g,%matplotlib inlinein a Notebook.show_ruler (bool) – if
True, add a ruler tool to the Matplotlib toolbar. Convenient to measure distances between peaks, etc.Warning
still experimental ! Try it, feedback welcome !
**kwargs (**dict) – kwargs forwarded as argument to plot (e.g: lineshape attributes:
lw=3, color='r')
- Returns:
line plot
- Return type:
line
Examples
Plot an
experimental_spectrum()in arbitrary units:s = experimental_spectrum(..., Iunit='mW/cm2/sr/nm') s.plot(Iunit='W/cm2/sr/cm-1')
See more examples in the plot Spectral quantities page.
See also
- plot_populations(what=None, nunit='', correct_for_abundance=False, **kwargs)[source]¶
Plots vib populations if given and format is valid.
- Parameters:
what (‘vib’, ‘rovib’, None) – if None plot everything
nunit (‘’, ‘cm-3’) – plot either in a fraction of vibrational levels, or a molecule number in in cm-3
correct_for_abundance (boolean) – if
True, multiplies each population by the isotopic abundance (as it is done during the calculation of emission integral)kwargs (**dict) – are forwarded to the plot
Examples
See also
- plot_slit(wunit=None, waveunit=None)[source]¶
Plot slit function that was applied to the Spectrum.
If dispersion was used (see
apply_slit()) the different slits are built again and plotted too (dotted).- Parameters:
wunit (
'nm','cm-1', orNone) – plot slit in wavelength or wavenumber. IfNone, use the unit the slit in which the slit function was given. DefaultNone- Returns:
fix, ax – figure and ax
- Return type:
matplotlib objects
Examples
s = radis.test_spectrum() s.apply_slit((0.4, 0.6), "nm") # add trapezoidal slit function s.plot_slit() # plot the slit function that was applied
See also
- populations[source]¶
populations of rovibrational levels used to compute partition functions of the spectrum
See also
get_populations(),plot_populations()
- print_conditions(**kwargs)[source]¶
Prints all physical / computational parameters.
- Parameters:
kwargs (dict) – refer to
print_conditions()
Examples
s = radis.test_spectrum() s.print_conditions()
You can also simply print the Spectrum object directly:
s = radis.test_spectrum() print(s)
Both syntaxes above will return (in radis==0.15):
# output >> Spectrum Name: CO-hitran-700K-#3680 Spectral Quantities ---------------------------------------- abscoeff [cm-1] (40,002 points) absorbance (40,002 points) emissivity_noslit (40,002 points) transmittance_noslit (40,002 points) radiance_noslit [mW/cm2/sr/cm-1] (40,002 points) Physical Conditions ---------------------------------------- Tgas 700 K Trot 700 K Tvib 700 K isotope 1,2,3 mole_fraction 0.1 molecule CO overpopulation None path_length 1 cm pressure_mbar 1013.25 mbar rot_distribution boltzmann self_absorption True state X thermal_equilibrium True vib_distribution boltzmann wavenum_max 2300.0000 cm-1 wavenum_min 1900.0000 cm-1 Computation Parameters ---------------------------------------- NwG 3 NwL 5 Tref 296 K add_at_used cython broadening_method voigt cutoff 1e-27 cm-1/(#.cm-2) dbformat hitran dbpath C:\Users\erwan\.radisdb\hitran\CO.hdf5 default_output_unit cm-1 diluents {'air': 0.9} folding_thresh 1e-06 include_neighbouring_lines True memory_mapping_engine auto neighbour_lines 0 cm-1 optimization simple parfuncfmt hapi parsum_mode full summation profiler {'spectrum_calculation': {'check_line_databank': ... pseudo_continuum_threshold 0 radis_version 0.14 sparse_ldm True spectral_points 40000.0 truncation 50 cm-1 waveunit cm-1 wstep 0.01 cm-1 zero_padding 40002 Config parameters ---------------------------------------- DEFAULT_DOWNLOAD_PATH ~/.radisdb GRIDPOINTS_PER_LINEWIDTH_ERROR_THRESHOLD 1 GRIDPOINTS_PER_LINEWIDTH_WARN_THRESHOLD 3 SPARSE_WAVERANGE auto Information ---------------------------------------- calculation_time 0.11549490000000162 s chunksize None db_use_cached True dxG 0.1375350788016573 dxL 0.20180288881201608 export_lines False export_populations None export_rovib_fraction True levelsfmt None lines_calculated 742 lines_cutoff 0 lines_in_continuum 0 load_energies False lvl_use_cached True parfuncpath None total_lines 742 warning_broadening_threshold 0.01 warning_linestrength_cutoff 0.01 wavenum_max_calc 2300.0000 cm-1 wavenum_min_calc 1900.0000 cm-1 ----------------------------------------
See also
- print_perf_profile(number_format='{:.3f}', precision=16)[source]¶
Prints Profiler output dictionary in a structured manner.
Example
Spectrum.print_perf_profile() # output >> spectrum_calculation 0.189s ████████████████ check_line_databank 0.000s check_non_eq_param 0.042s ███ fetch_energy_5 0.015s █ calc_weight_trans 0.008s reinitialize 0.002s copy_database 0.000s memory_usage_warning 0.002s reset_population 0.000s calc_noneq_population 0.041s ███ part_function 0.035s ██ population 0.006s scaled_non_eq_linestrength 0.005s map_part_func 0.001s corrected_population_se 0.003s calc_emission_integral 0.006s applied_linestrength_cutoff 0.002s calc_lineshift 0.001s calc_hwhm 0.007s generate_wavenumber_arrays 0.001s calc_line_broadening 0.074s ██████ precompute_LDM_lineshapes 0.012s LDM_Initialized_vectors 0.000s LDM_closest_matching_line 0.001s LDM_Distribute_lines 0.001s LDM_convolve 0.060s █████ others 0.001s calc_other_spectral_quan 0.003s generate_spectrum_obj 0.000s others -0.016s- Other Parameters:
precision (int, optional) – total number of blocks. Default 16.
See also
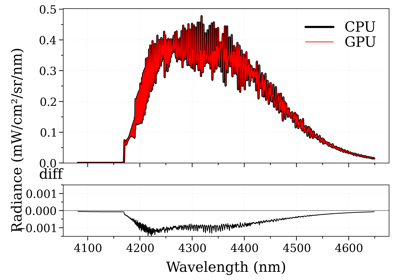
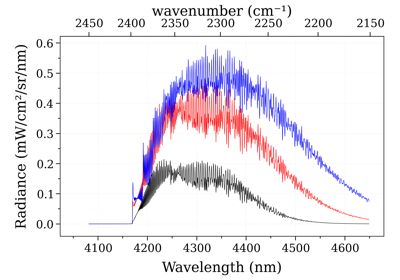
- recalc_gpu(var='abscoeff', wunit='default', Iunit='default', pressure=None, Tgas=None, mole_fraction=None, path_length=None, slit_function=None)[source]¶
Recalculate the spectrum based on new input parameters. Can only be called for spectrum objects produced by
eq_spectrum_gpu(). This method is used internally byeq_spectrum_gpu_interactive(). Parameters may be passed as arguments, or updated directly inconditions, after which spectrum.recalc_gpu() may be called without passing arguments.- Parameters:
var (variable (
absorbance,transmittance,transmittance_noslit,xsection, etc.)) – For full list seeget_vars(). IfNone, plot the first thing in the Spectrum. DefaultNone.wunit (
'nm','cm-1','nm_vac'orNone) – wavelength in air ('nm'), wavenumber ('cm-1'), or wavelength in vacuum ('nm_vac'). IfNone,'wavespace'must be defined inconditions.Iunit (unit for variable) – if
default, default unit for quantityvaris used. for radiance, one can use per wavelength (~W/m2/sr/nm) or per wavenumber (~W/m2/sr/cm-1) unitspressure (float, optional) – Pressure in [bar]
Tgas (float, optional) – Equilibrium temperature in [K]
mole_fraction (float, optional) – Mole fraction
path_length (float, optional) – Absroption length in [cm]
slit_function (float, optional) – Slit FWHM in [cm-1]
- Returns:
new_y – The recalculated array specified by the
varkeyword.- Return type:
numpy.ndarray[np.fltoat32]
Examples
s = sf.eq_spectrum_gpu( Tgas=1100.0, # K pressure=1, # bar mole_fraction=0.8, path_length=0.2, # cm ) I = [] for T in [300.0, 1000.0, 2000.0]: I.append(s.recalc_gpu('radiance', Tgas=T)
- resample(w_new, unit='same', out_of_bounds='nan', energy_threshold='default', print_conservation=False, inplace=True, if_conflict_drop=None, **kwargs)[source]¶
Resample spectrum over a new wavelength/wavenumber range.
Warning
This may result in information loss. Resampling is done with oversampling and spline interpolation. These parameters can be adjusted, and energy conservation ensured with the appropriate parameters.
To minimize information loss, always resample the high-resolution spectrum over the low-resolution spectrum, i.e.
s_highres.resample(s_lowres)
Fills with
'nan'or transparent medium (transmittance 1, radiance 0) when out of bound (seeout_of_bounds)- Parameters:
w_new (array, or Spectrum) – new wavespace to resample the spectrum on. Must be inclosed in the current wavespace (we won’t extrapolate) One can also give a Spectrum directly:
s1.resample(s2.get_wavenumber()) s1.resample(s2) # also valid
unit (
'same','nm','cm-1','nm_vac') – unit of new wavespace. It'same'it is assumed to be the current waveunit. Default'same'. The spectrum waveunit is changed to this unit after resampling (i.e: a spectrum calculated and stored incm-1but resampled innmwill be stored innmfrom now on). If'nm', wavelength in air. If'nm_vac', wavelength in vacuum.out_of_bounds (
'transparent','nan','error') – what to do if resampling is out of bounds.'transparent': fills with transparent medium. ‘nan’: fill with nan.'error': raises an error. Default'nan'medium (
'air','vacuum', or'default') – in which medium is the new waverange is calculated if it is given in ‘nm’. Ignored if unit=’cm-1’
- Other Parameters:
energy_threshold (float or
Noneor'default') -- if energy conservation (integrals on the intersecting range) is above this threshold, raise an error. If ``None, dont check for energy conservation. If'default', look up the value inradis.config[“RESAMPLING_TOLERANCE_THRESHOLD”] Default'default'print_conservation (boolean) – if
True, prints energy conservation. DefaultFalse.inplace (boolean) – if
True, modifies the Spectrum object directly. Else, returns a copy. DefaultTrue.**kwargs (**dict) – all other arguments are sent to
radis.misc.signal.resample()
- Returns:
Spectrum – object has been modified anyway.
- Return type:
resampled Spectrum object. If using
inplace=True, the Spectrum
Examples
Convert a Spectrum in wavenumber to wavelengths:
s_nm = s.resample(s.get_wavelength(), "nm", inplace=False)
- resample_even(energy_threshold=0.005, print_conservation=False, inplace=True)[source]¶
Resample spectrum over the same waverange, but evenly spaced.
Warning
This may result in information loss. Resampling is done with oversampling and spline interpolation. These parameters can be adjusted, and energy conservation ensured with the appropriate parameters.
- Parameters:
energy_threshold (float or
None) – if energy conservation (integrals on the intersecting range) is above this threshold, raise an error. IfNone, dont check for energy conservation Default 5e-3 (0.5%)print_conservation (boolean) – if
True, prints energy conservation. DefaultFalse.inplace (boolean) – if
True, modifies the Spectrum object directly. Else, returns a copy. DefaultTrue.**kwargs (**dict) – all other arguments are sent to
radis.misc.signal.resample()
- Returns:
Spectrum – object has been modified anyway.
- Return type:
resampled Spectrum object. If using
inplace=True, the Spectrum
Examples
- rescale_mole_fraction(new_mole_fraction, old_mole_fraction=None, inplace=True, ignore_warnings=False, force=False, verbose=True)[source]¶
Update spectrum with new molar fraction Convoluted values (with slit) are dropped in the process.
- Parameters:
new_mole_fraction (float) – new mole fraction
old_mole_fraction (float, or None) – if None, current mole fraction (conditions[‘mole_fraction’]) is used
- Other Parameters:
inplace (boolean) – if
True, modifies the Spectrum object directly. Else, returns a copy. DefaultTrue.force (boolean) – if False, won’t allow rescaling to 0 (not to loose information). Default
False
- Returns:
s – Cropped Spectrum. If
inplace=True, Spectrum has been updated directly anyway. Allows chaining- Return type:
Examples
s.rescale_mole_fraction(0.2)
Notes
Implementation:
similar to rescale_path_length() but we have to scale abscoeff & emisscoeff Note that this is valid only for small changes in mole fractions. Then, the change in line broadening becomes significant
See also
- rescale_path_length(new_path_length, old_path_length=None, inplace=True, force=False)[source]¶
Rescale spectrum to new path length. Starts from absorption coefficient and emission coefficient, and solves the RTE again for the new path length Convoluted values (with slit) are dropped in the process.
- Parameters:
new_path_length (float) – new path length
old_path_length (float, or None) – if None, current path length (conditions[‘path_length’]) is used
- Other Parameters:
inplace (boolean) – if
True, modifies the Spectrum object directly. Else, returns a copy. DefaultTrue.force (boolean) – if False, won’t allow rescaling to 0 (not to loose information). Default
False
- Returns:
s – Cropped Spectrum. If
inplace=True, Spectrum has been updated directly anyway. Allows chaining- Return type:
Examples
for path in [0.1, 10, 100]: s.rescale_path_length(10, inplace=False).plot(nfig='same')
Additionally, you can also use astropy units in the input arguments, for example:
# preparing a test spectrum : import radis s = radis.test_spectrum() # rescaling : import astropy.units as u s.rescale_path_length(1 * u.km).plot()
Notes
Implementation:
To deal with all the input cases, we first make a list of what has to be recomputed, and what has to be recalculated
See also
- savetxt(filename, var, wunit='default', Iunit='default')[source]¶
Export spectral quantity var to filename.
- (note that by doing this you will loose additional information, such
as the calculation conditions or the units. You better save a Spectrum object under a .spec file with
store()and load it afterwards withload_spec())
- Parameters:
filename (str) – file name
var (str) – which spectral variable ot export
- Other Parameters:
wunit, Iunit, medium (str) – see
get()for more information
Notes
Export variable as:
np.savetxt(filename, np.vstack(self.get(var, wunit=wunit, Iunit=Iunit, medium=medium)).T, header=header)
See also
- sort(inplace=True)[source]¶
Sort the Spectrum by wavelength / wavenumber.
- Parameters:
inplace (bool, optional) – if
True, modifies the Spectrum object directly. The default isTrue.- Returns:
s – sorted Spectrum. If
inplace=True, Spectrum has been updated directly anyway. Allows chaining.- Return type:
Examples
- store(path, discard=['lines', 'populations'], compress=True, add_info=None, add_date=None, if_exists_then='error', verbose=True)[source]¶
Save a Spectrum object in JSON format. Object can be recovered with
load_spec(). If many Spectrum are saved in a same folder you can view their properties with theSpecDatabasestructure.- Parameters:
path (path to folder (database) or file) – if a folder, file is saved to database and name is generated automatically. if a file name, then Spectrum is saved to this file and the later formatting options dont apply
file (str) – explicitly give a filename to save
compress (boolean) – if
False, save under text format, readable with any editor. ifTrue, saves under binary format. Faster and takes less space. If2, removes all quantities that can be regenerated with s.update(), e.g, transmittance if abscoeff and path length are given, radiance if emisscoeff and abscoeff are given in non-optically thin case, etc. DefaultTrue.add_info (list) – append these parameters and their values if they are in conditions example:
add_info = ['Tvib', 'Trot']
discard (list of str) – parameters to exclude, for instance to save some memory for instance Default [
lines,populations]: retrieved Spectrum looses theline_survey()ability, andplot_populations()(but it saves tons of memory!)if_exists_then (
'increment','replace','error','ignore') – what to do if file already exists. If increment an incremental digit is added. If replace file is replaced (yeah). If'ignore'no file is created. If error (or anything else) an error is raised. Defaulterror
- Return type:
Returns filename used
Notes
If many spectra are stored in a folder, it may be time to set up a
SpecDatabasestructure to easily see all Spectrum conditions and get Spectrum that suits specific parameters.Examples
Store a spectrum in compressed mode, regenerate quantities after loading:
from radis import load_spec s.store('test.spec', compress=True) # s is a Spectrum s2 = load_spec('test.spec') s2.update() # regenerate missing quantities
Examples using
radis.spectrum.spectrum.Spectrum.store¶See also
- take(var, copy_lines=False, copy_arrays=True)[source]¶
- Parameters:
var (str) – spectral quantity (
'absorbance','transmittance','xsection'etc.)- Other Parameters:
copy_lines (bool) – if
True, exports.lines. DefaultFalsecopy_arrays (bool) – if
False, returned array’s quantity is a pointer to the original Spectrum. Faster, but warning, changing them will then change the original Spectrum. DefaultTrue
- Returns:
s – same Spectrum with only the
varspectral quantity- Return type:
Examples
Use
taketo chain other commandss.take('radiance').normalize().plot()
- to_hdf5(file, engine='pytables')[source]¶
Stores the Spectrum under HDF5 format. Uses
spec2hdf()Examples
s.to_hdf5('spec01.h5')
See also
- to_json(*args, **kwargs)[source]¶
Alias to Spectrum.store(compress=False).
See Spectrum.
store()for documentation
- to_pandas(copy=True)[source]¶
Convert a Spectrum to a Pandas DataFrame
- Returns:
pd.DataFrame – units are stored in attributes
df.attrs- Return type:
pandas DataFrame where columns are spectral arrays, and
Notes
Pandas does not support units yet. pint-pandas is an advanced project but not fully working. See discussion in https://github.com/pandas-dev/pandas/issues/10349
For the moment, we store units as metadata
- to_specutils(var=None, wunit='default', Iunit='default')[source]¶
Convert a
radisSpectrumobject tospecutilsspecutils.spectra.spectrum1d.Spectrum1D- Parameters:
var (‘radiance’, ‘radiance_noslit’, etc.) – which spectral array to convert. If
Noneand only one spectral array is defined, use it.wunit (
'nm','cm','nm_vac'.) – wavespace unit: wavelength in air ('nm'), wavenumber ('cm-1'), or wavelength in vacuum ('nm_vac'). if"default", default unit for waveunit is used. Seeget_waveunit().Note
specutilshandles wavelengths in vacuum only. If using'nm'wavelengths will be converted to wavelengths in vacuum. We recommend using'nm_air'or'nm_vac'to avoid any confusion.Iunit (unit for variable
var) – if"default", default unit for quantityvaris used. See theunitsattribute. Forvar="radiance", one can use per wavelength (~ ‘W/m2/sr/nm’) or per wavenumber (~ ‘W/m2/sr/cm-1’) unitsNote
nanthat may have been added on the wings of the spectra if a slit has been applied are removed usingtrim_nanparameter ofget(). The waverange in the 1DSpectrum object may therefore be cropped.
Examples
spectrum = s.to_specutils()
Add uncertainties by reading a spectrum noise region with
SpectralRegionandnoise_region_uncertainty()from specutils import SpectralRegion from specutils.manipulation import noise_region_uncertainty noise_region = SpectralRegion(2012 / u.cm, 2009 / u.cm) spectrum = noise_region_uncertainty(spectrum, noise_region)
Find lines out of the noise using
find_lines_threshold()from specutils.fitting import find_lines_threshold lines = find_lines_threshold(spectrum)
See also
- trim(inplace=True)[source]¶
Remove
nancommon to all arrays on each side of the Spectrum.Returns a smaller Spectrum (inplace or not).
- Returns:
s – directly anyway. Allows chaining.
- Return type:
Spectrum : trimmed Spectrum. If
inplace=True, Spectrum has been updated
- update(quantity='all', optically_thin='default', verbose=True)[source]¶
Calculate missing quantities: ex: if path_length and emisscoeff are given, recalculate radiance_noslit.
- Parameters:
spec (Spectrum)
quantity (str) – name of the spectral quantity to recompute. If ‘same’, only the quantities in the Spectrum are recomputed. If ‘all’, then all quantities that can be derived are recomputed. Default ‘all’.
optically_thin (True, False, or ‘default’) – determines whether to calculate radiance with or without self absorption. If ‘default’, the value is determined from the self_absorption key in Spectrum.conditions. If not given, False is taken. Default ‘default’ Also updates the self_absorption value in conditions (creates it if doesnt exist
Examples
Initialize a spectrum from the absorption coefficient, retrieve the transmittance or the emission coefficient :
s = Spectrum.from_array([1900. , 1900.01, 1900.02, 1900.03, 1900.04, 1900.05, 1900.06, 1900.07, 1900.08, 1900.09], [2.71414065e-06, 2.88341489e-06, 3.06942277e-06, 3.27445689e-06, 3.50121831e-06, 3.75290756e-06, 4.03334037e-06, 4.34709612e-06, 4.69971017e-06, 5.09792551e-06], 'abscoeff', wunit='cm-1', Iunit='cm-1', conditions={'path_length':1, # cm 'thermal_equilibrium':True, 'Tgas':700, # K }) s.update('transmittance_noslit') s.update('emisscoeff')
See also