See populations of computed levels¶
Spectrum methods gives you access to the populations of levels used in a nonequilibrium spectrum :
get_populations()provides the populations
of all levels of the molecule, used to compute the nonequilibrium partition function
linesreturns all informations
of the visible lines, in particular the populations of the upper and lower levels of absorbing/emitting lines in the spectrum.
from astropy import units as u
We first compute a noneq CO spectrum.
Notice the keys export_lines=True and export_populations=True
from radis import calc_spectrum
s = calc_spectrum(
1900 / u.cm,
2300 / u.cm,
molecule="CO",
isotope="1",
pressure=1.01325 * u.bar,
Tvib=3000 * u.K,
Trot=1000 * u.K,
mole_fraction=0.1,
path_length=1 * u.cm,
databank="hitran", # or 'hitemp', 'geisa', 'exomol'
export_lines=True,
export_populations="rovib",
)
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/io/hitran.py:170: AccuracyWarning:
All columns are not downloaded currently, please use cache = 'regen' and extra_params='all' to download all columns.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: HighTemperatureWarning:
HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
Calculating Non-Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000.0 K
Trot 1000.0 K
Tvib 3000.0 K
isotope 1
mole_fraction 0.1
molecule CO
path_length 1.0 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2300.0000 cm-1
wavenum_min 1900.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/.radisdb/hitran/CO.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm auto
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding -1
----------------------------------------
Fetching Evib & Erot.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: NegativeEnergiesWarning:
There are negative rotational energies in the database
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: PerformanceWarning:
'gu' was recomputed although 'gp' already in DataFrame. All values are equal
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: PerformanceWarning:
'object' type column found in database, calculations and memory usage would be faster with a numeric type. Possible solution is to not use 'save_memory' and convert the columns to dtype.
... sorting lines by vibrational bands
... lines sorted in 0.0s
0.13s - Spectrum calculated
Below we plot the populations
We could also have used plot_populations() directly
pops = s.get_populations("CO")["rovib"]
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/spectrum/spectrum.py:2410: UserWarning:
Populations valid for partition function calculation but sometimes NOT for spectra calculations, e.g. CO2.
See help on how to use 's.lines.query' instead, for instance in https://github.com/radis/radis/issues/508.
Turn off warning with 'show_warning=False'
Plot populations :
Here we plot population fraction vs rotational number of the 2nd vibrational level v==2,
for all levels as well as for levels of lines visible in the spectrum
import matplotlib.pyplot as plt
pops.query("v==2").plot(
"j",
"n",
label="populations used to compute\n partition functions",
style=".-",
markersize=2,
)
s.lines.query("vl==2").plot(
"jl",
"nl",
ax=plt.gca(),
label="populations of visible\n absorbing lines",
kind="scatter",
color="r",
)
plt.xlim((0, 80))
plt.legend()
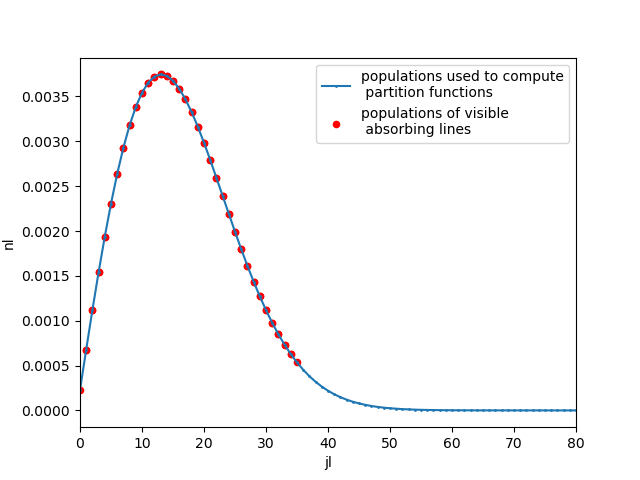
<matplotlib.legend.Legend object at 0x7fbdf29cd5a0>
Print all levels information
print(f"{len(pops)} levels ")
print(pops)
print(pops.columns)
7766 levels
v j E ... n Qrot nrot
0 0 0 0.000000 ... 1.754208e-03 362.691791 0.002757
1 0 1 3.845033 ... 5.233590e-03 362.691791 0.008226
2 0 2 11.534953 ... 8.626674e-03 362.691791 0.013559
3 0 3 23.069466 ... 1.187857e-02 362.691791 0.018670
4 0 4 38.448131 ... 1.493823e-02 362.691791 0.023479
... .. .. ... ... ... ... ...
7761 48 38 89299.135499 ... 2.237048e-21 360.424333 0.003612
7762 48 39 89447.643402 ... 1.853601e-21 360.424333 0.002993
7763 48 40 89599.882150 ... 1.526677e-21 360.424333 0.002465
7764 48 41 89755.845909 ... 1.249931e-21 360.424333 0.002018
7765 48 42 89915.528699 ... 1.017299e-21 360.424333 0.001643
[7766 rows x 13 columns]
Index(['v', 'j', 'E', 'Evib', 'viblvl', 'gj', 'gvib', 'grot', 'Erot', 'nvib',
'n', 'Qrot', 'nrot'],
dtype='object')
Print visible lines information :
print(f"{len(s.lines)} visible lines in the spectrum")
print(s.lines)
print(s.lines.columns)
274 visible lines in the spectrum
wav int A ... hwhm_voigt hwhm_lorentz hwhm_gauss
0 1900.341956 3.539000e-29 12.45 ... 0.016713 0.015650 0.004067
1 1902.414114 2.295000e-31 24.91 ... 0.017474 0.016455 0.004072
2 1905.778633 9.041000e-29 12.54 ... 0.016872 0.015814 0.004079
3 1907.676341 5.311000e-31 25.11 ... 0.017672 0.016659 0.004083
4 1911.187699 2.268000e-28 12.64 ... 0.017028 0.015973 0.004091
.. ... ... ... ... ... ... ...
269 2291.548195 1.124000e-28 21.70 ... 0.017153 0.015650 0.004905
270 2293.336784 4.416000e-29 21.77 ... 0.017008 0.015491 0.004909
271 2295.082667 1.703000e-29 21.83 ... 0.016901 0.015372 0.004912
272 2296.785698 6.450000e-30 21.90 ... 0.016752 0.015208 0.004916
273 2298.445736 2.400000e-30 21.97 ... 0.016609 0.015049 0.004919
[274 rows x 55 columns]
Index(['wav', 'int', 'A', 'airbrd', 'selbrd', 'El', 'Tdpair', 'Pshft', 'ierr',
'iref', 'line_mixing_flag', 'gp', 'gpp', 'Fu', 'branch', 'jl', 'syml',
'Fl', 'vu', 'vl', 'ju', 'Eu', 'Evibl', 'Evibu', 'Erotu', 'Erotl', 'gju',
'gjl', 'grotu', 'grotl', 'gvibu', 'gvibl', 'gu', 'gl', 'Rs2', 'Blu',
'Bul', 'Aul', 'viblvl_l', 'viblvl_u', 'band', 'Qrotl', 'Qrotu',
'nu_vib', 'nl_vib', 'nu_rot', 'nl_rot', 'nu', 'nl', 'S', 'Ei',
'shiftwav', 'hwhm_voigt', 'hwhm_lorentz', 'hwhm_gauss'],
dtype='object')
We can also look at unique levels; for instance sorting by quantum numbers v, J
of the lower level : vl, jl
print(
len(s.lines.groupby(["vl", "jl"])),
" visible absorbing levels (i.e. unique 'vl', 'jl' ",
)
141 visible absorbing levels (i.e. unique 'vl', 'jl'
line_survey() is also a convenient way to explore populations and other line
parameters, for instance
s.line_survey(overlay="absorbance", barwidth=0.001, lineinfo="all")

Total running time of the script: (0 minutes 0.448 seconds)



