HITRAN spectra¶
The absorption coefficient of all HITRAN species (see MOLECULES_LIST_EQUILIBRIUM)
is calculated in plot_all_hitran_spectra.py
at 300 K, 1 atm for the first isotope:
1
'H2O': Water (spectrum)2
'CO2': Carbon Dioxide (spectrum)3
'O3': Ozone (spectrum)4
'N2O': Nitrogen oxide (spectrum)5
'CO': Carbon Monoxide (spectrum)6
'CH4': Methane (spectrum)7
'O2': Oxygen8
'NO': Nitric Oxide (spectrum)9
'SO2': Sulfur Dioxide (spectrum)10
'NO2': Nitrogen Dioxide (spectrum)11
'NH3': Ammonia (spectrum)12
'HNO3': Nitric Acid (spectrum)13
'OH': Hydroxyl (spectrum)14
'HF': Hydrogen Fluoride (spectrum)15
'HCl': Hydrogen Chloride (spectrum)16
'HBr': Hydrogen Bromide (spectrum)17
'HI': Hydrogen Iodide (spectrum)18
'ClO': Chlorine Monoxide (spectrum)19
'OCS': Carbonyl Sulfide (spectrum)20
'H2CO': Formaldehyde (spectrum)21
'HOCl': Hypochlorous Acid (spectrum)22
'N2': Nitrogen23
'HCN': Hydrogen Cyanide24
'CH3Cl': Methyl Chloride (spectrum)25
'H2O2': Hydrogen Peroxide (spectrum)26
'C2H2': Acetylene (spectrum)27
'C2H6': Ethane (spectrum)28
'PH3': Phosphine (spectrum)29
'COF2': Carbonyl Fluoride (spectrum)30
'SF6': Sulfur Hexafluoride31
'H2S': Hydrogen Sulfide (spectrum)32
'HCOOH': Formic Acid (spectrum)33
'HO2': Hydroperoxyl (spectrum)34
'O': Oxygen Atom35
'ClONO2': Chlorine Nitrate36
'NO+': Nitric Oxide Cation (spectrum)37
'HOBr': Hypobromous Acid38
'C2H4': Ethylene39
'CH3OH': Methanol40
'CH3Br': Methyl Bromide41
'CH3CN': Acetonitrile42
'CF4': CFC-1443
'C4H2': Diacetylene44
'HC3N': Cyanoacetylene45
'H2': Hydrogen46
'CS': Carbon Monosulfide47
'SO3': Sulfur trioxide48
'C2N2': Cyanogen49
'COCl2': Phosgene
The code to calculate each molecule is shown below:
1. H2O¶
1
'H2O': Water absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='H2O', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
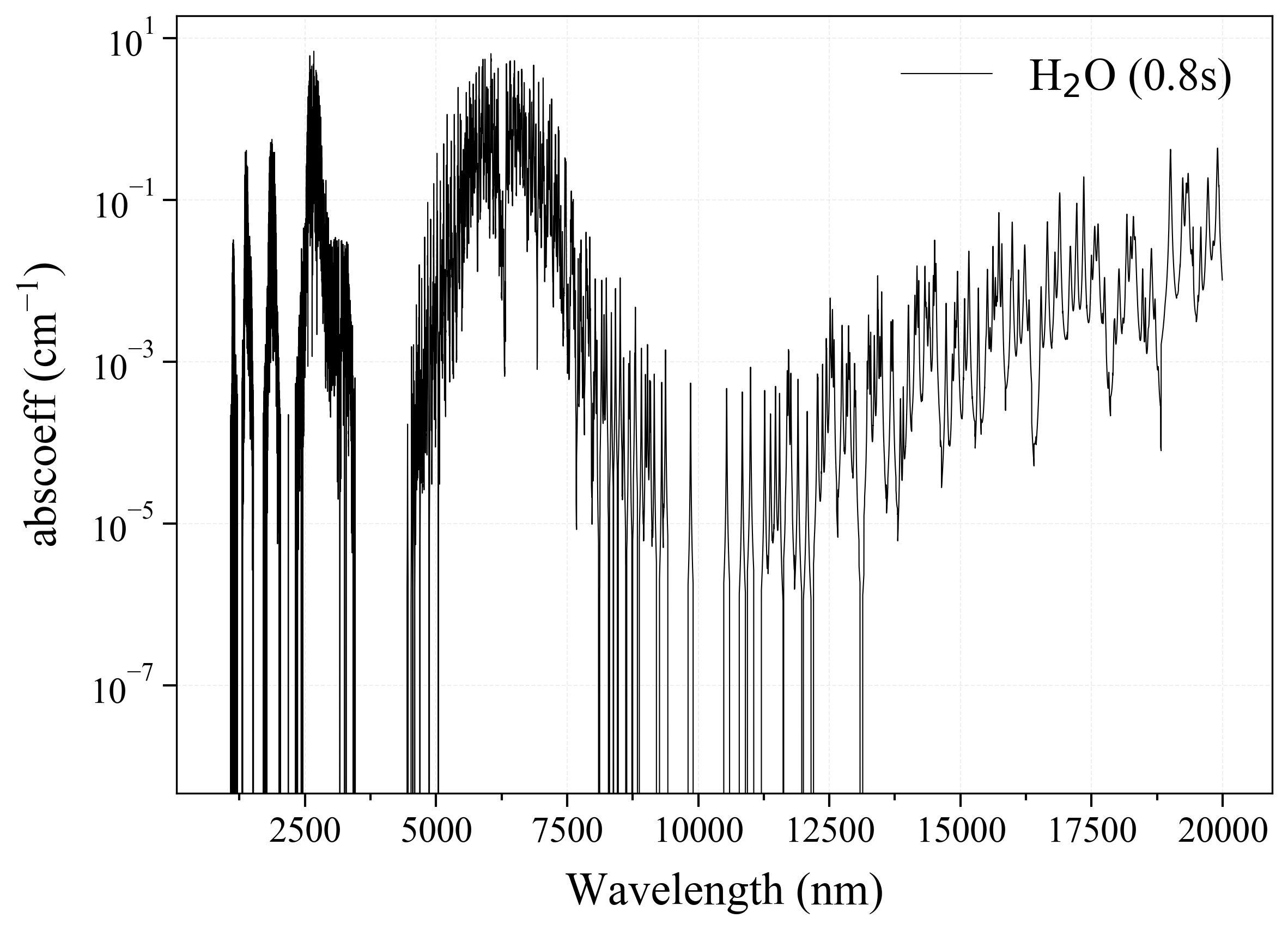
2. CO2¶
2
'CO2': Carbon Dioxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='CO2', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
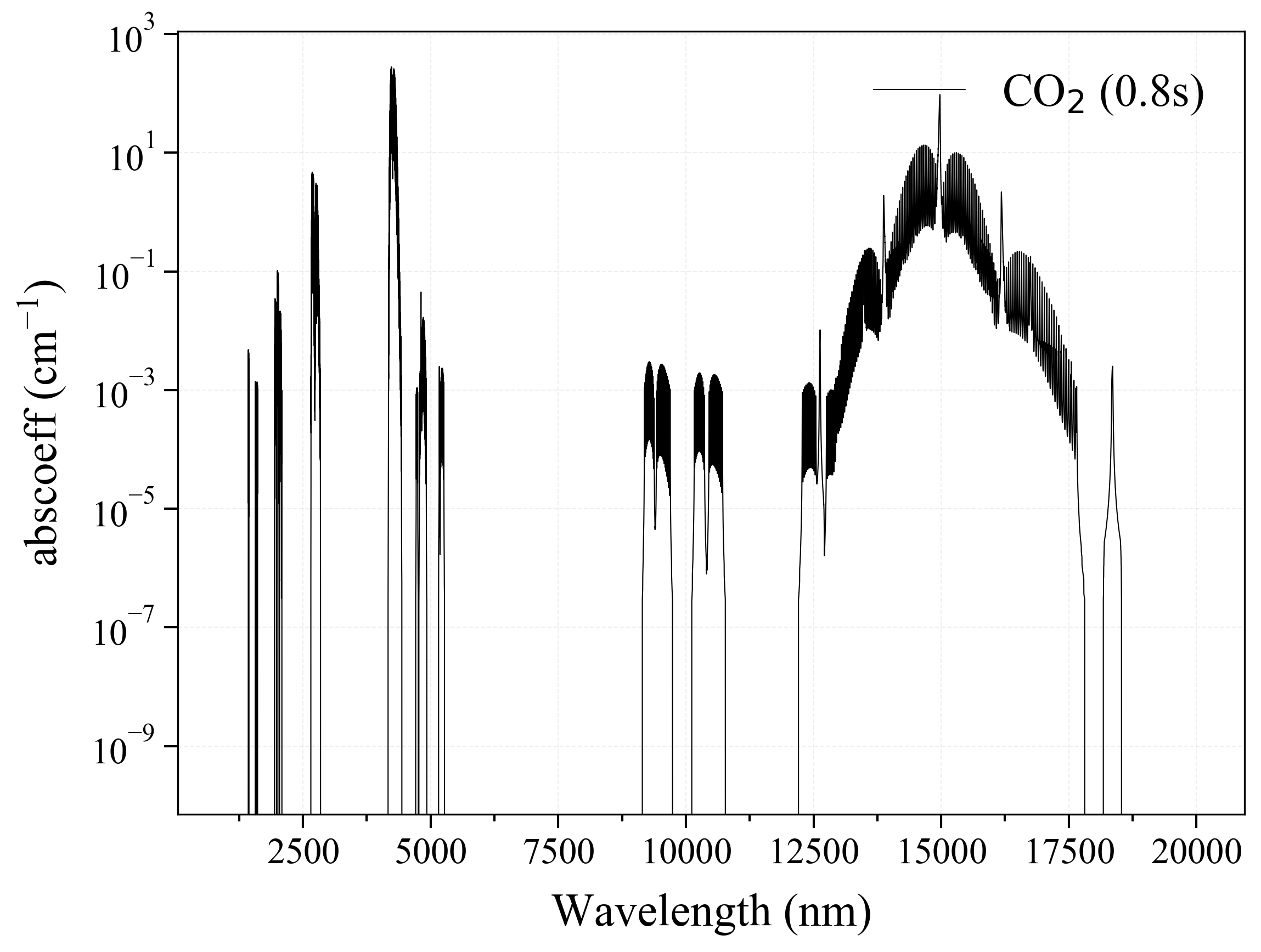
3. O3 =====-
3
'O3': Ozone absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='O3', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')

4. N2O¶
4
'N2O': Nitrogen oxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='N2O', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
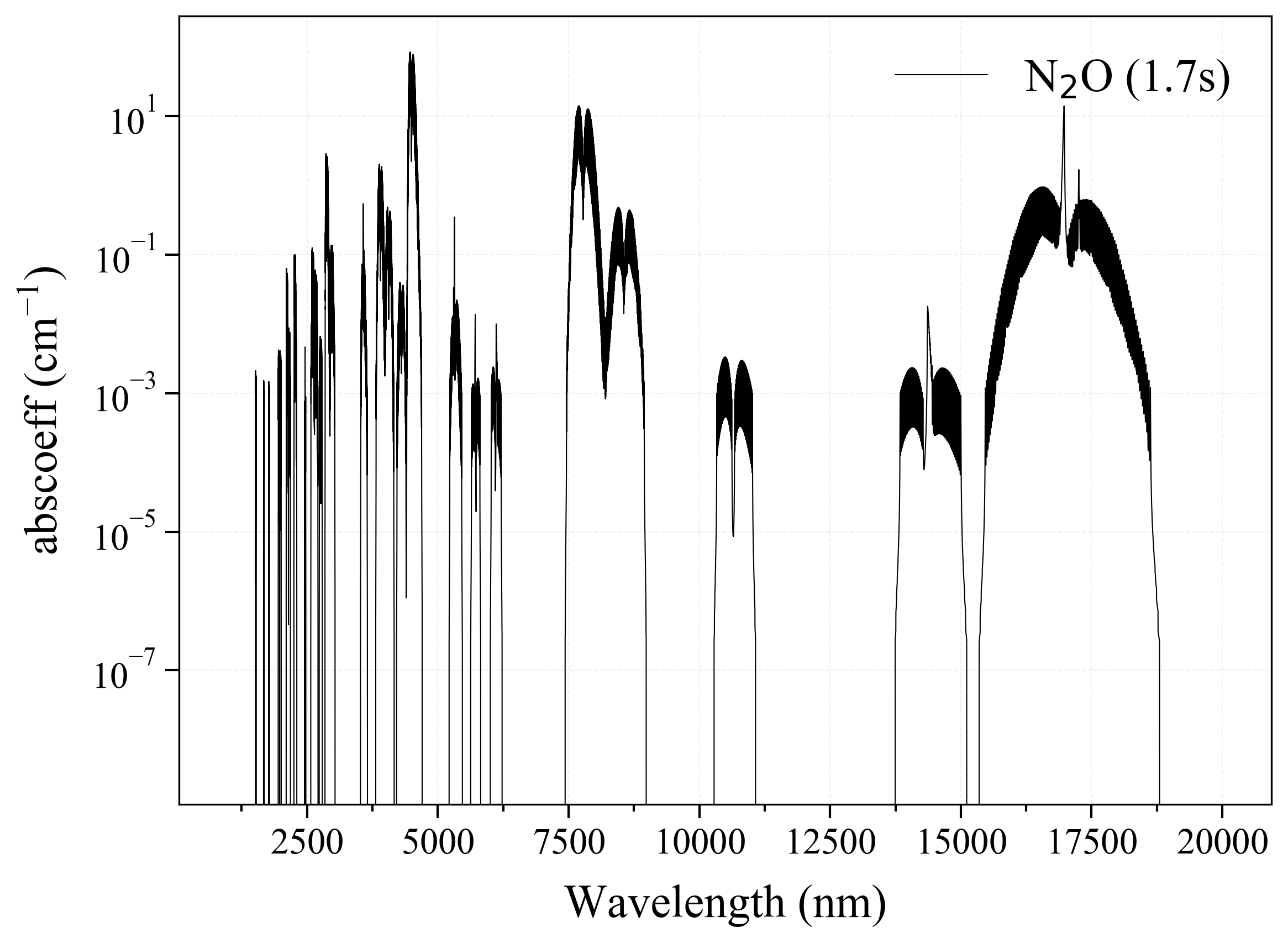
5. CO¶
5
'CO': Carbon Monoxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='CO', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')

6. CH4¶
6
'CH4': Methane absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='CH4', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
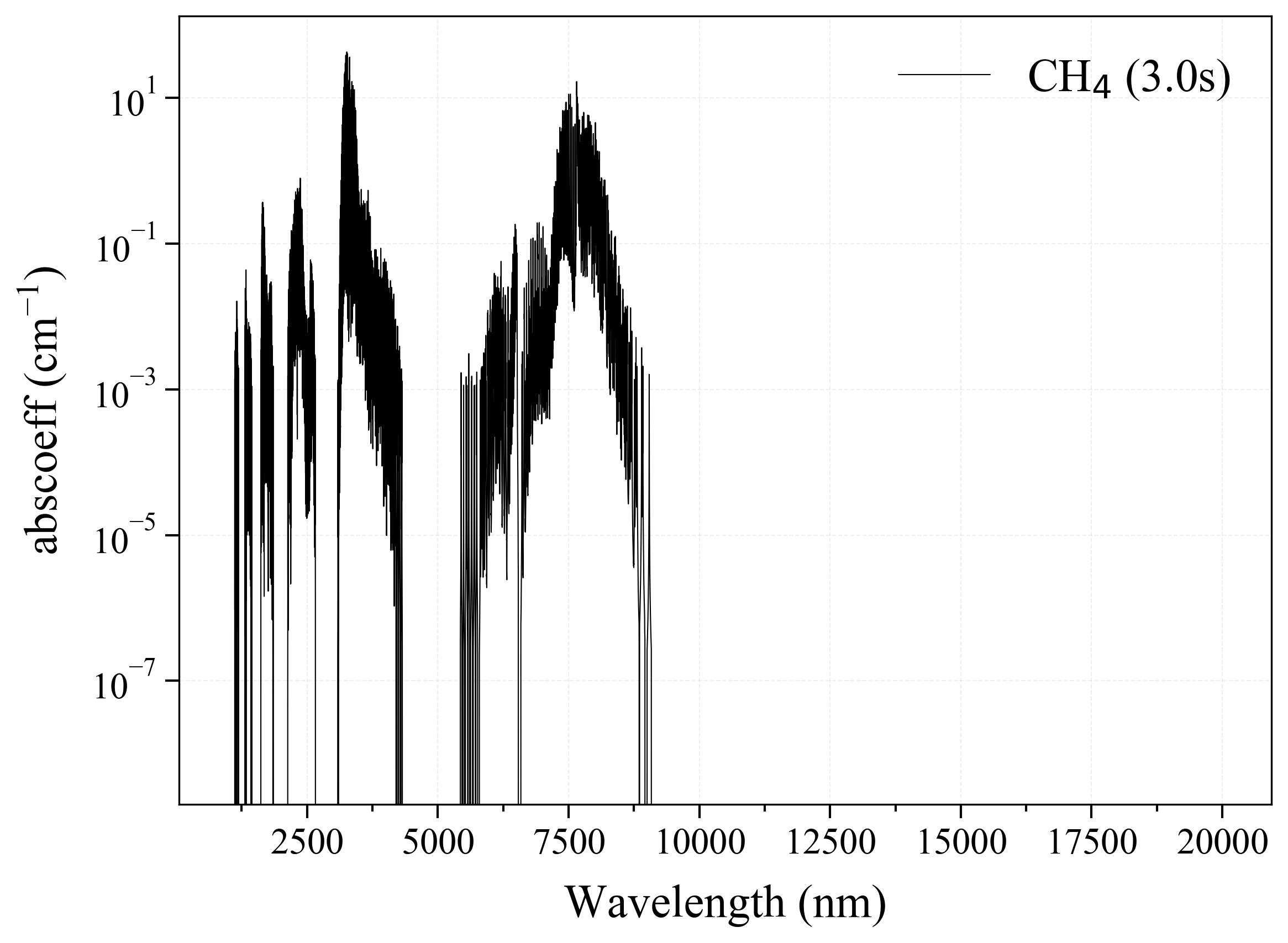
7. O2 =====-
7
'O2': Oxygen absorption coefficient (opacity) at 300 K : no lines forisotope='1'(symmetric!)
8. NO¶
8
'NO': Nitric Oxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='NO', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
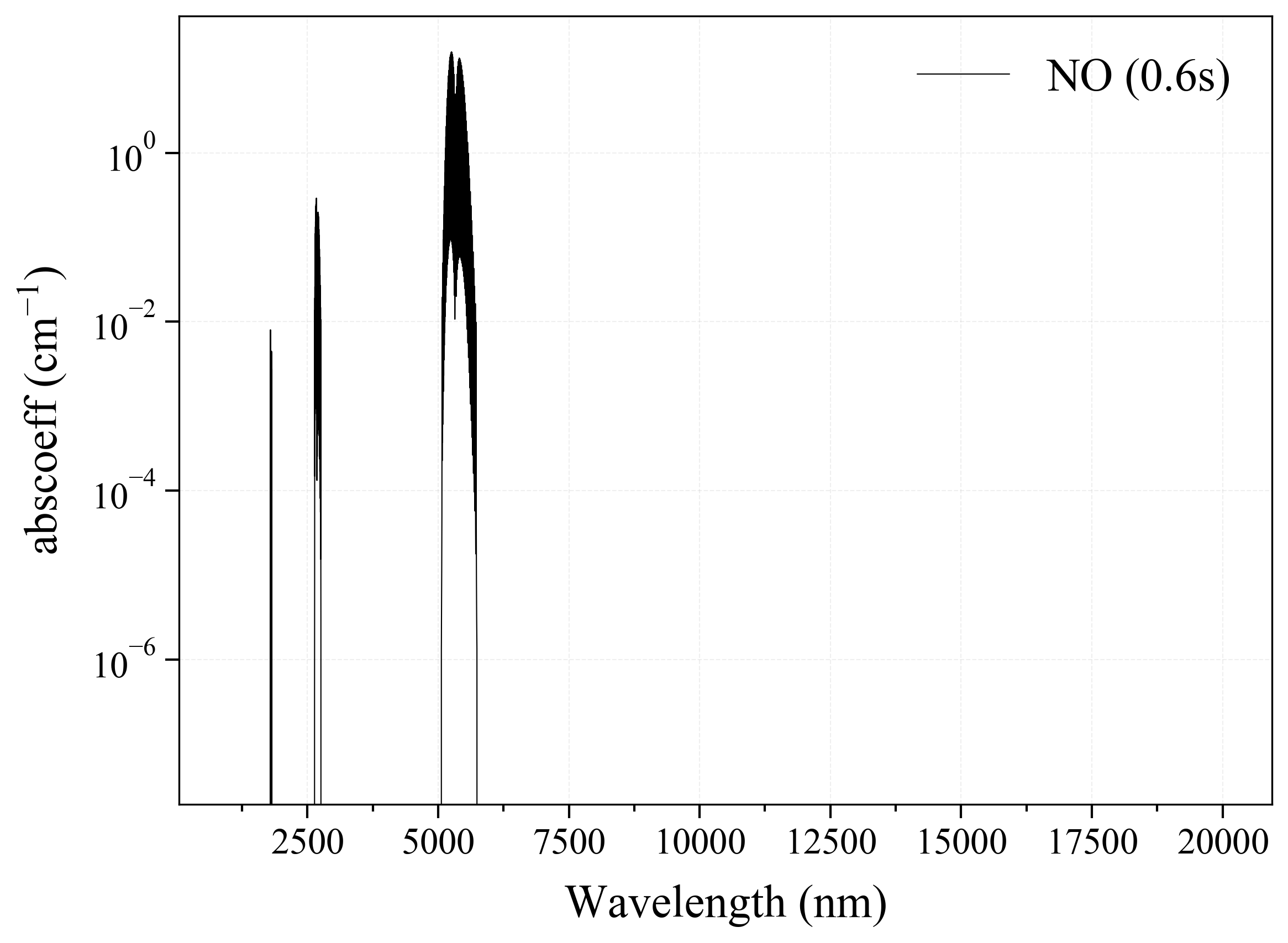
9. SO2¶
9
'SO2': Sulfur Dioxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='SO2', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
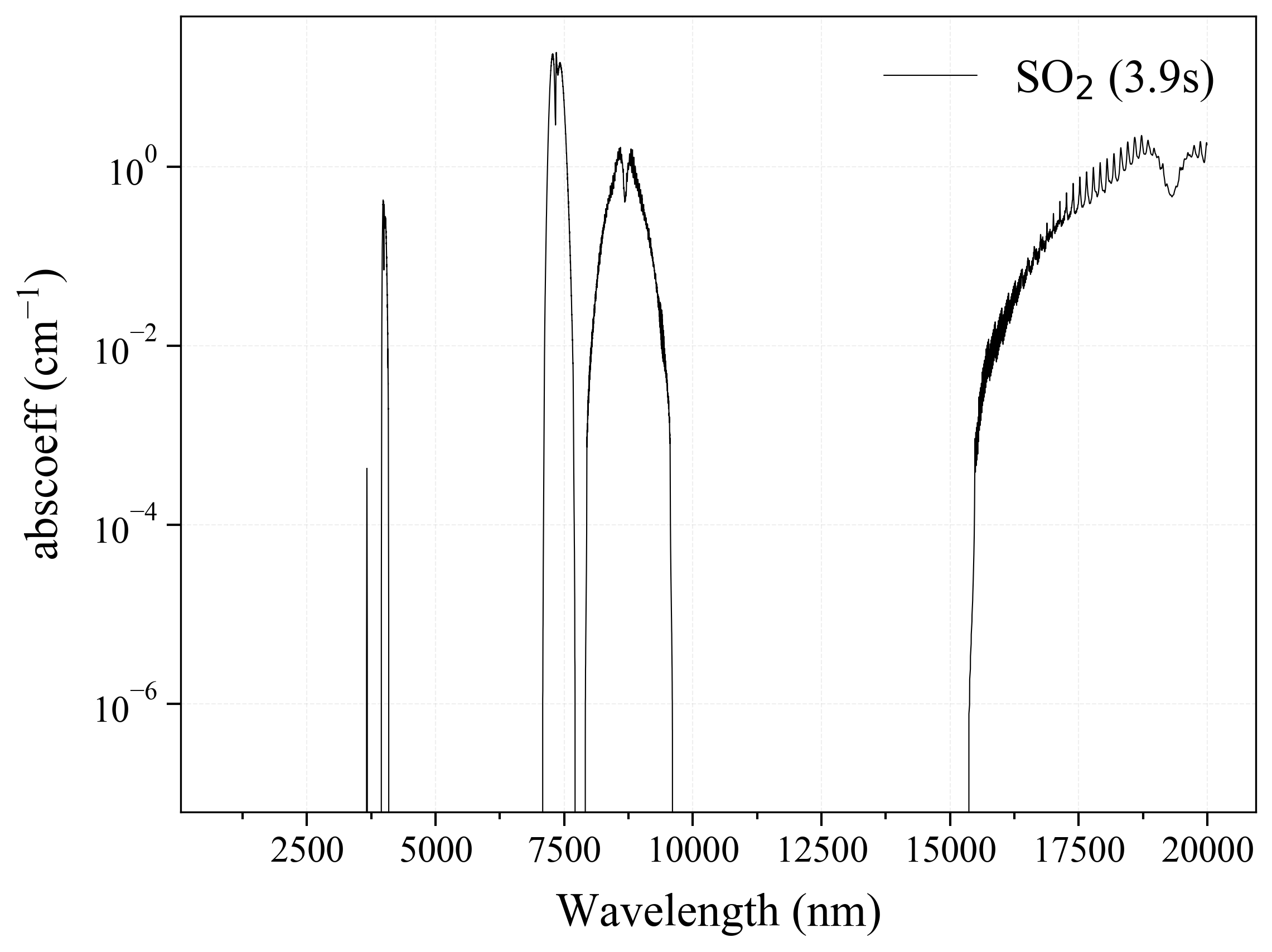
10. NO2¶
10
'NO2': Nitrogen Dioxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='NO2', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
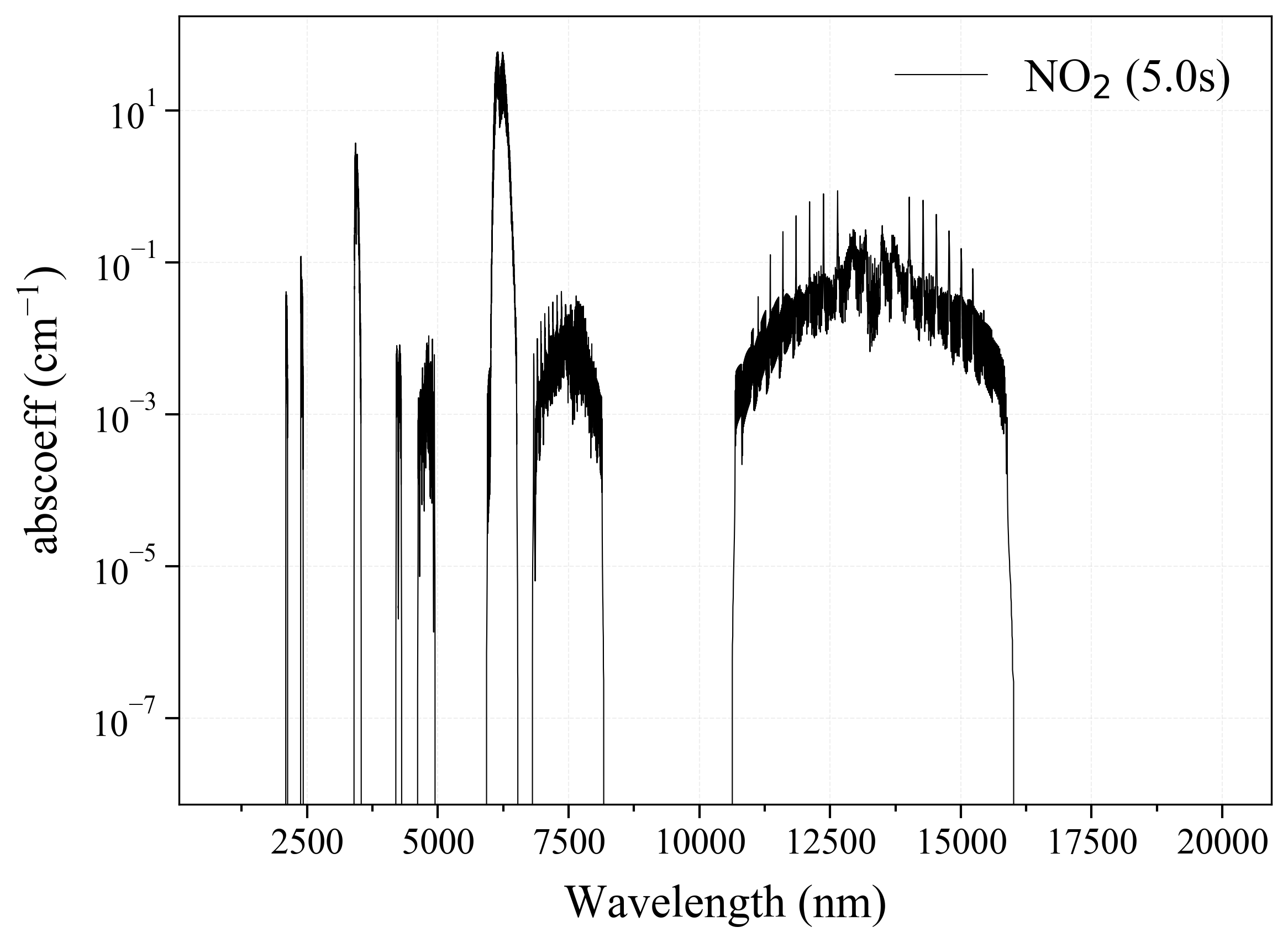
11. NH3¶
11
'NH3': Ammonia absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='NH3', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
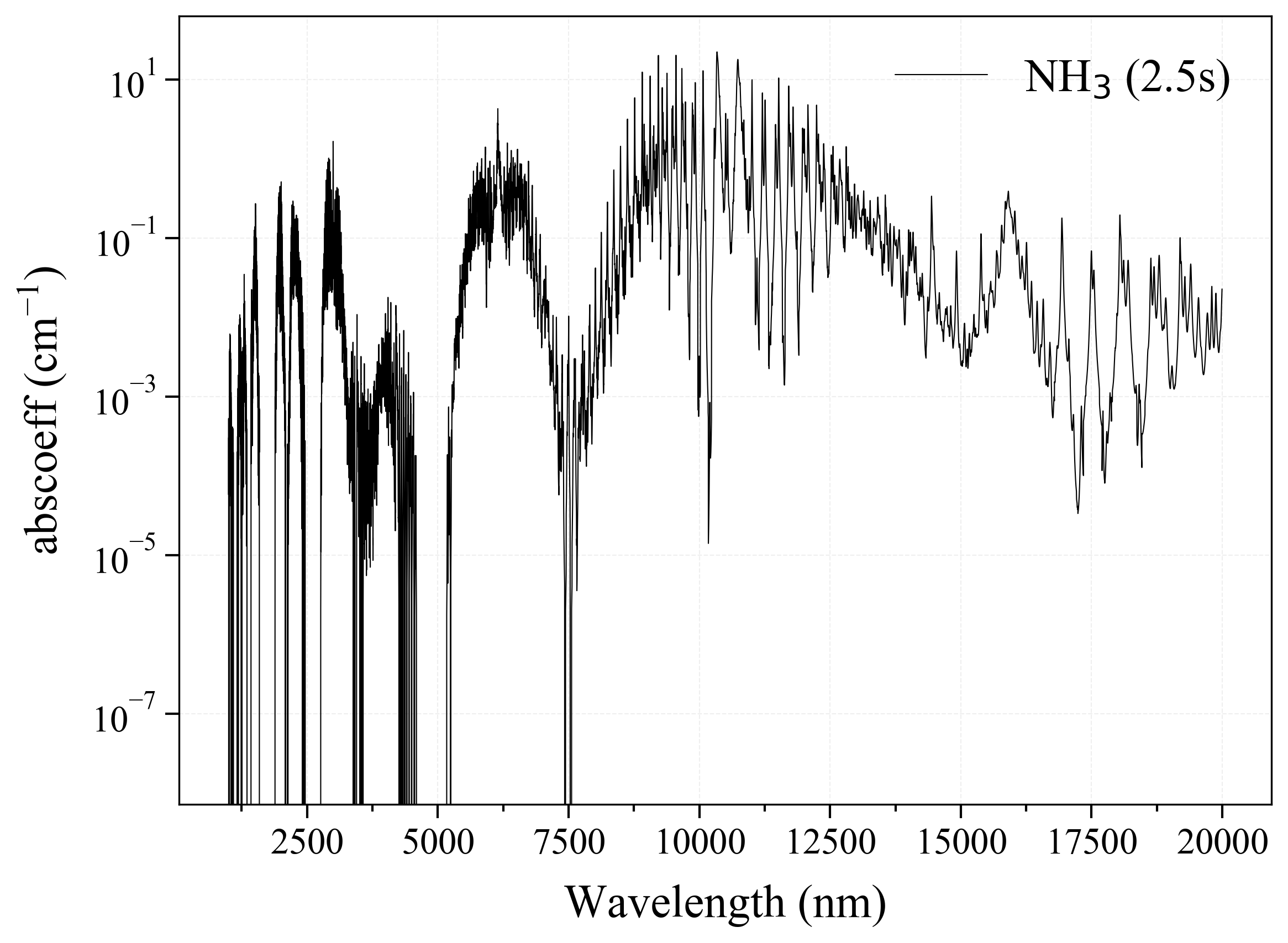
12. HNO3¶
12
'HNO3': Nitric Acid absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HNO3', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')

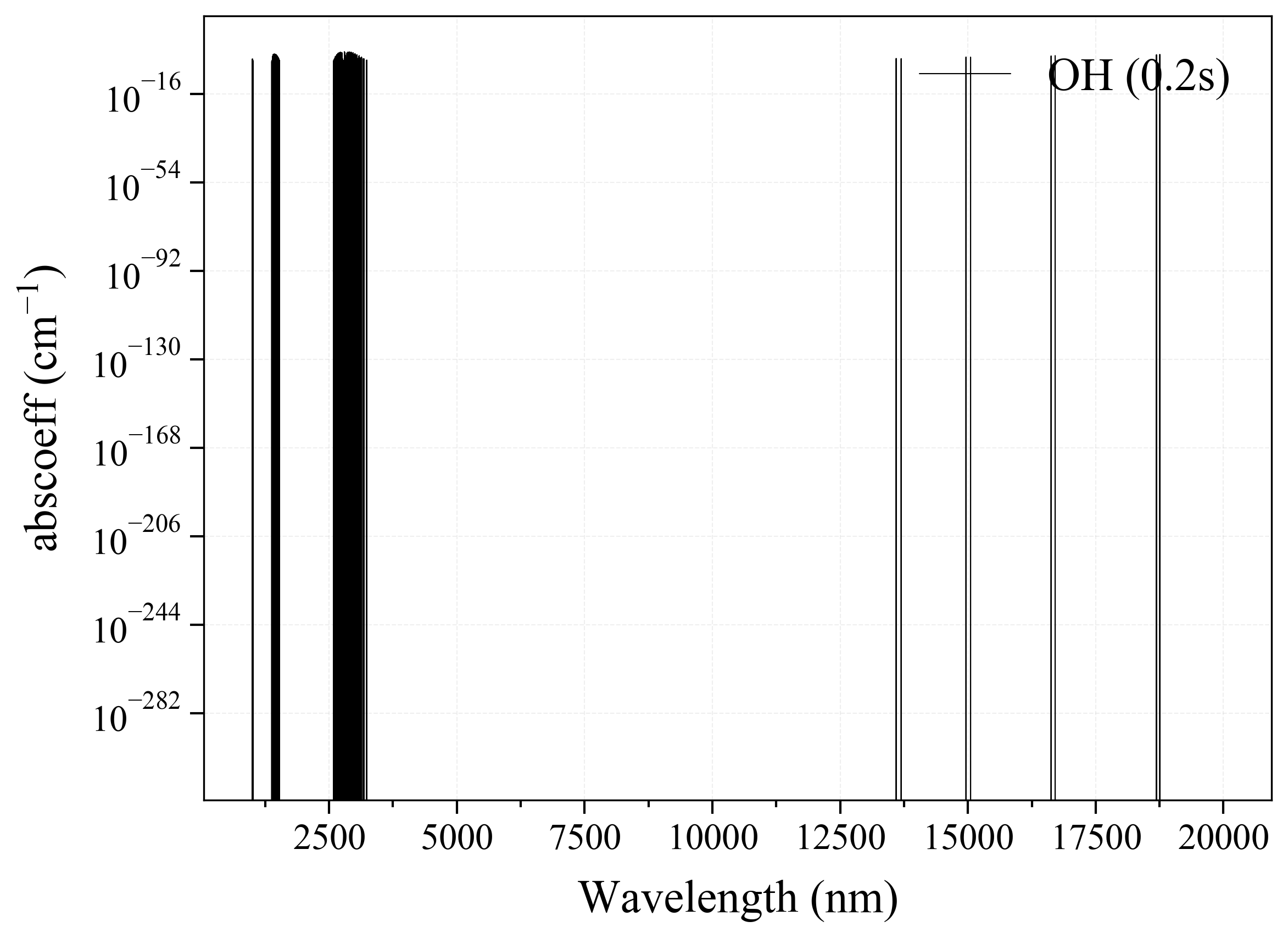
13. OH¶
13
'OH': Hydroxyl absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='OH', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
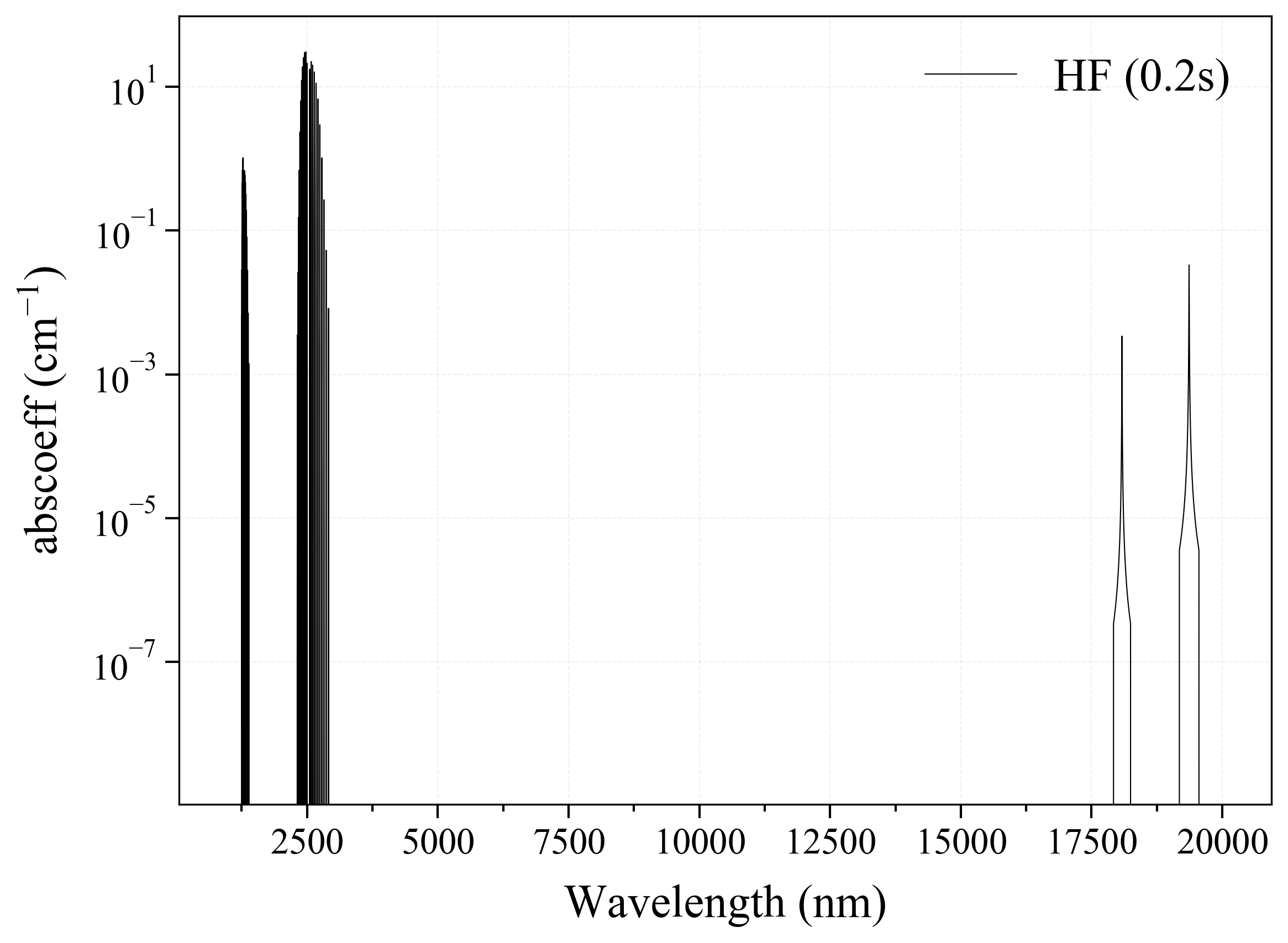
14. HF¶
14
'HF': Hydrogen Fluoride absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HF', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
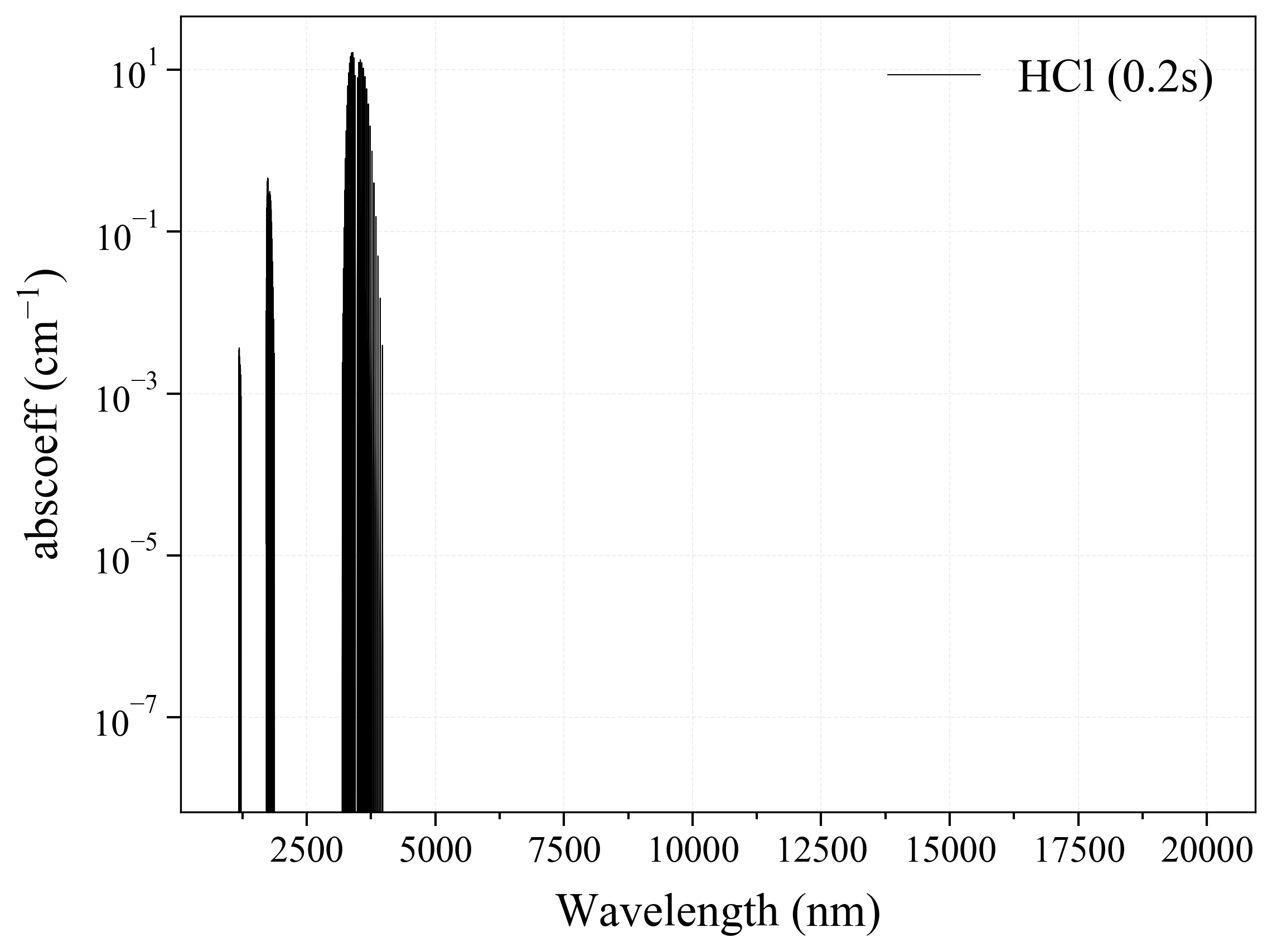
15. HCl¶
15
'HCl': Hydrogen Chloride absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HCl', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
16. HBr¶
16
'HBr': Hydrogen Bromide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HBr', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')

17. HI¶
17
'HI': Hydrogen Iodide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HI', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')

18. ClO¶
18
'ClO': Chlorine Monoxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='ClO', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')

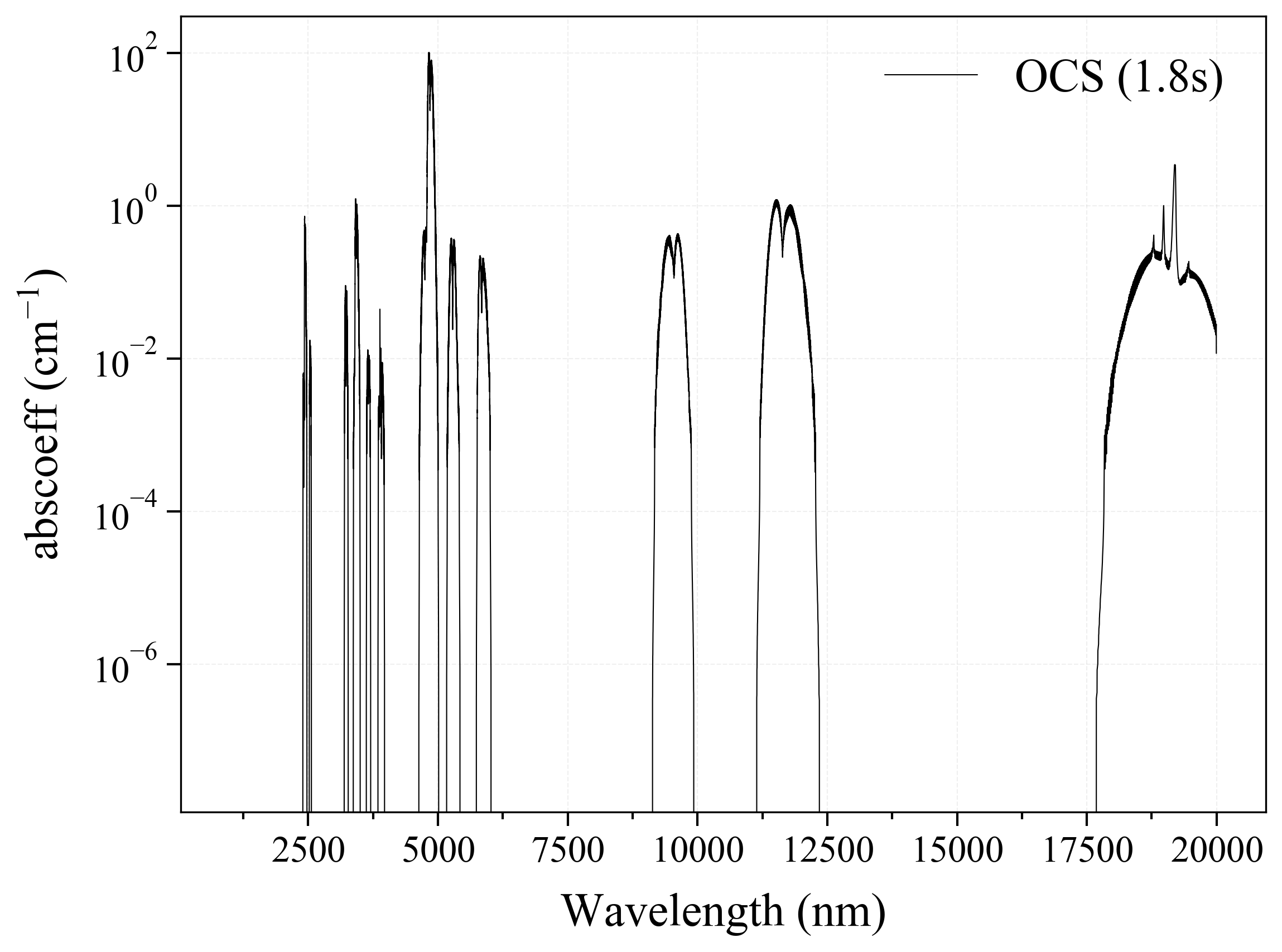
19. OCS¶
19
'OCS': Carbonyl Sulfide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='OCS', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
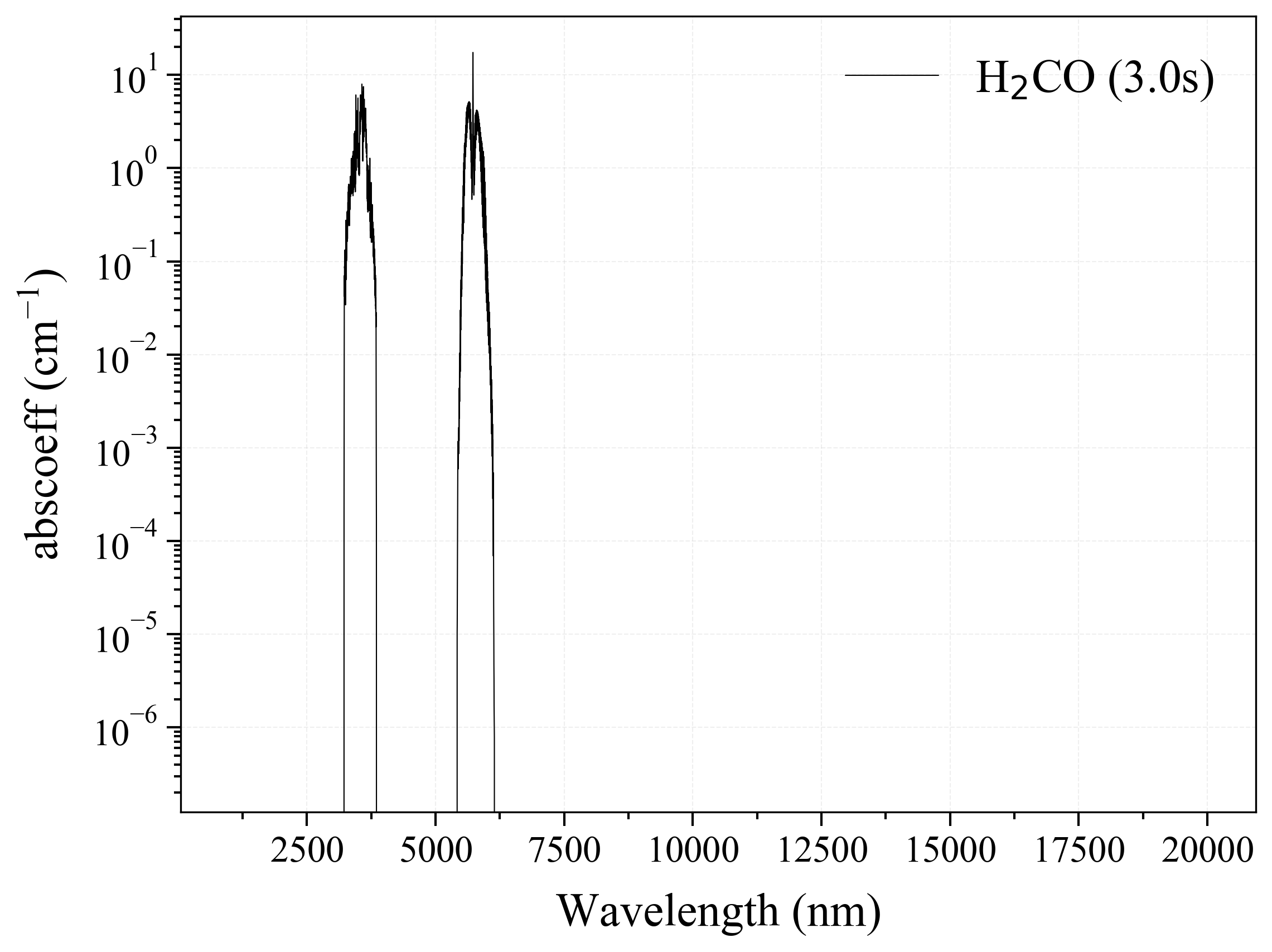
20. H2CO¶
20
'H2CO': Formaldehyde absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='H2CO', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
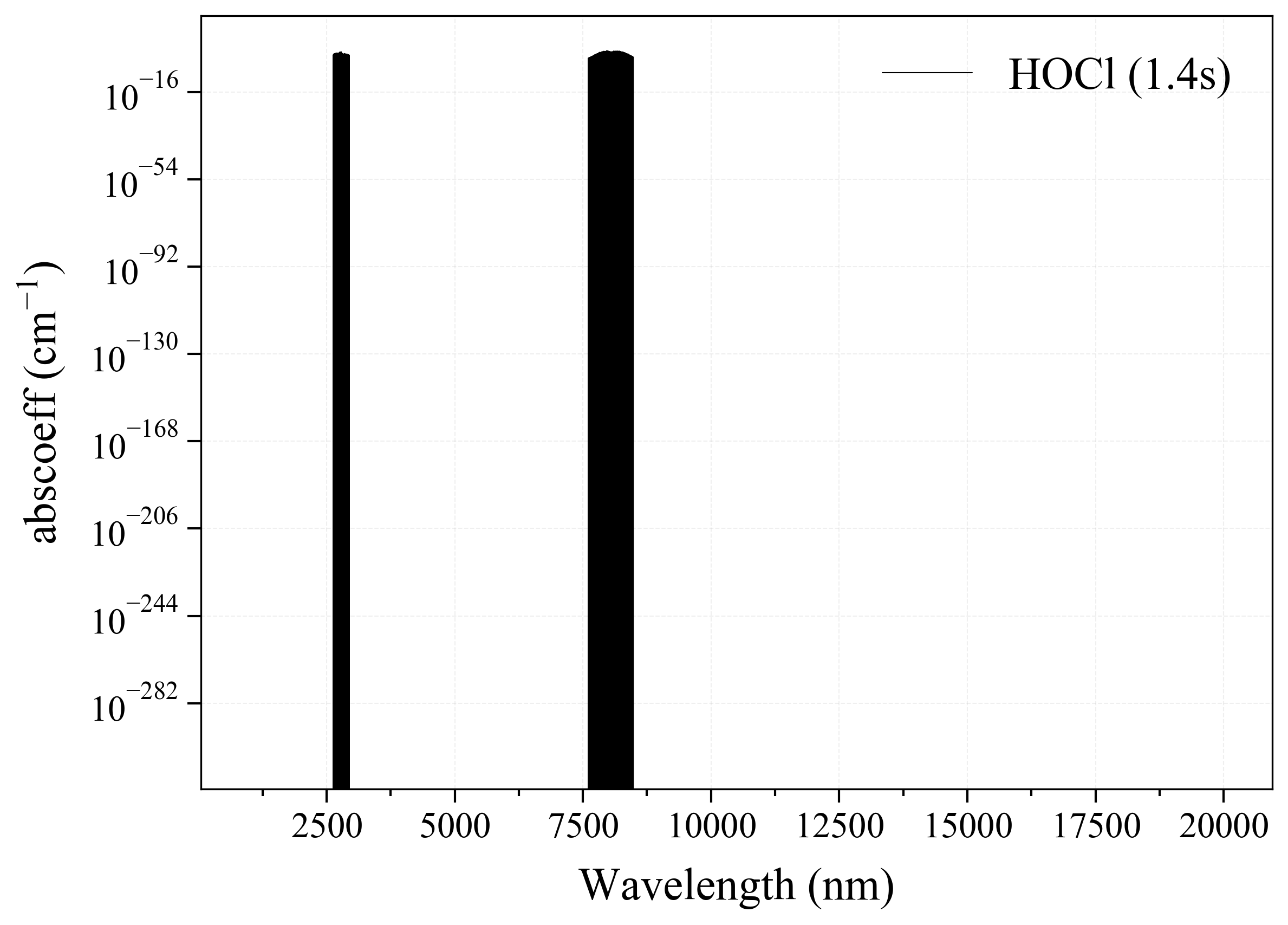
21. HOCl¶
21
'HOCl': Hypochlorous Acid absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HOCl', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
22. N2¶
22
'N2': Nitrogen absorption coefficient (opacity) at 300 K : no lines forisotope='1'(symmetric!)
23. HCN¶
- 23
'HCN'Hydrogen Cyanide absorption coefficient (opacity) at 300 Knot calculated.
- 23
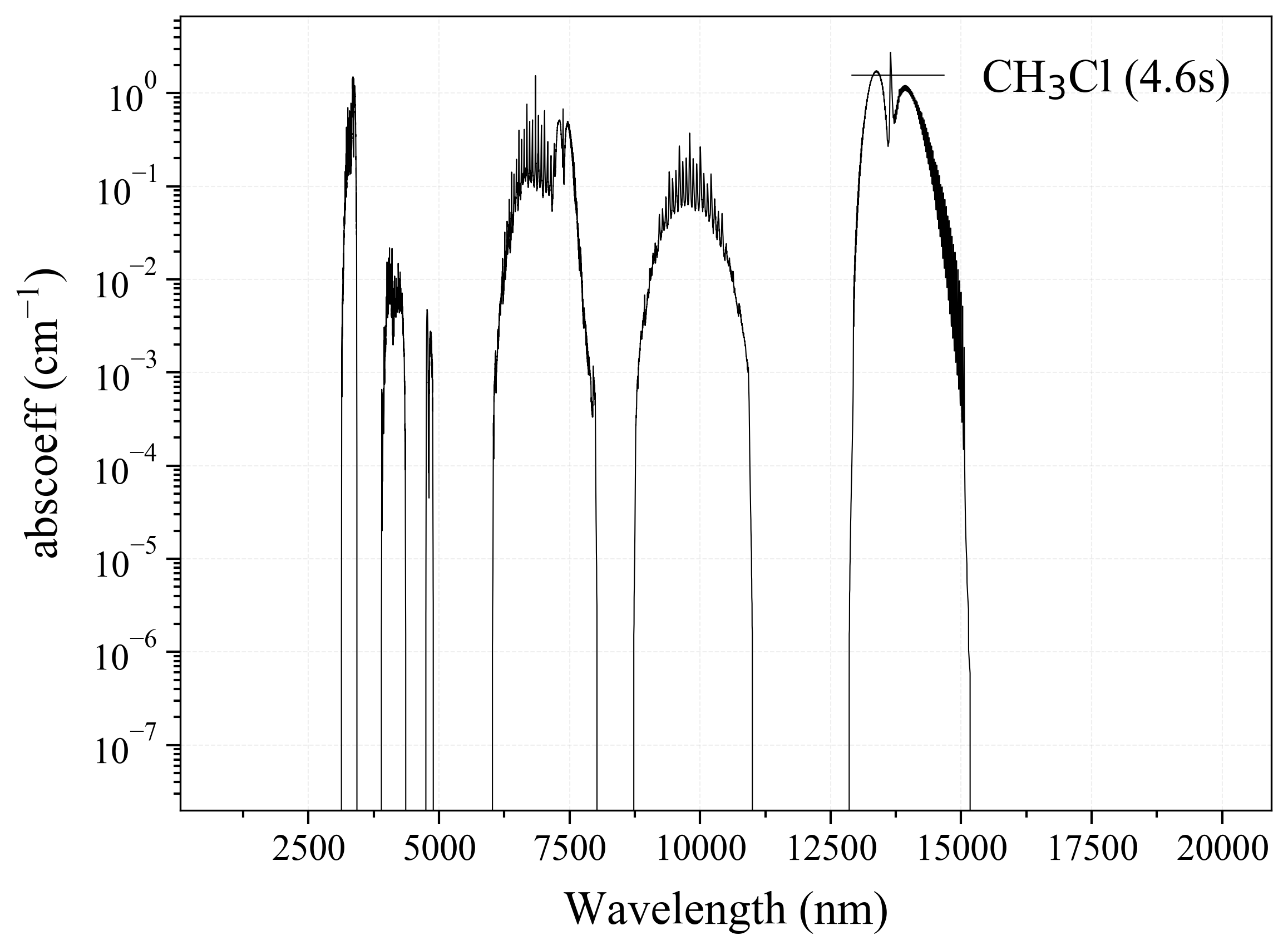
24. CH4Cl¶
24
'CH3Cl': Methyl Chloride absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='CH3Cl', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
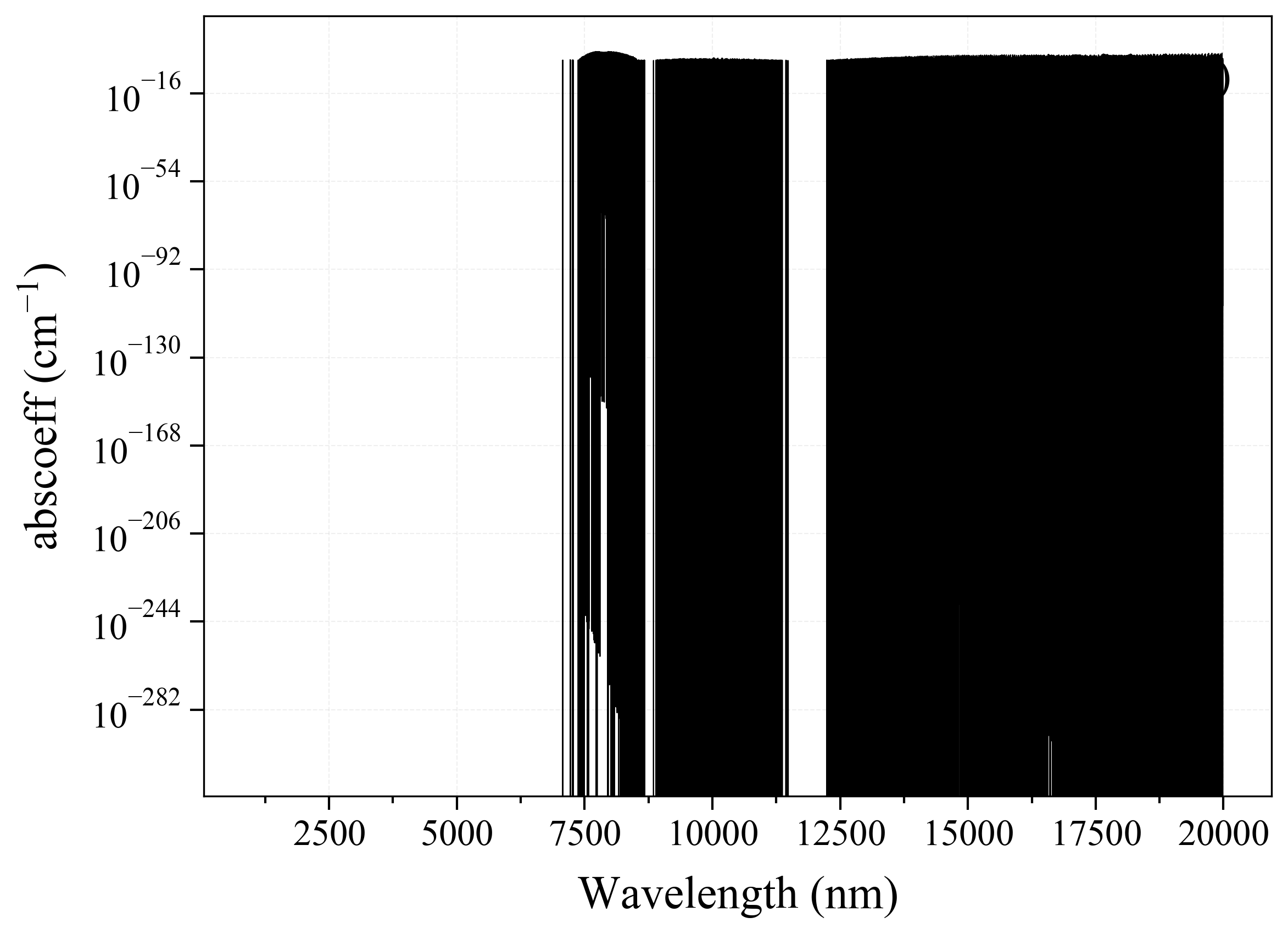
25. H2O2¶
25
'H2O2': Hydrogen Peroxide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='H2O2', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
26. C2H2¶
26
'C2H2': Acetylene absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='C2H2', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')

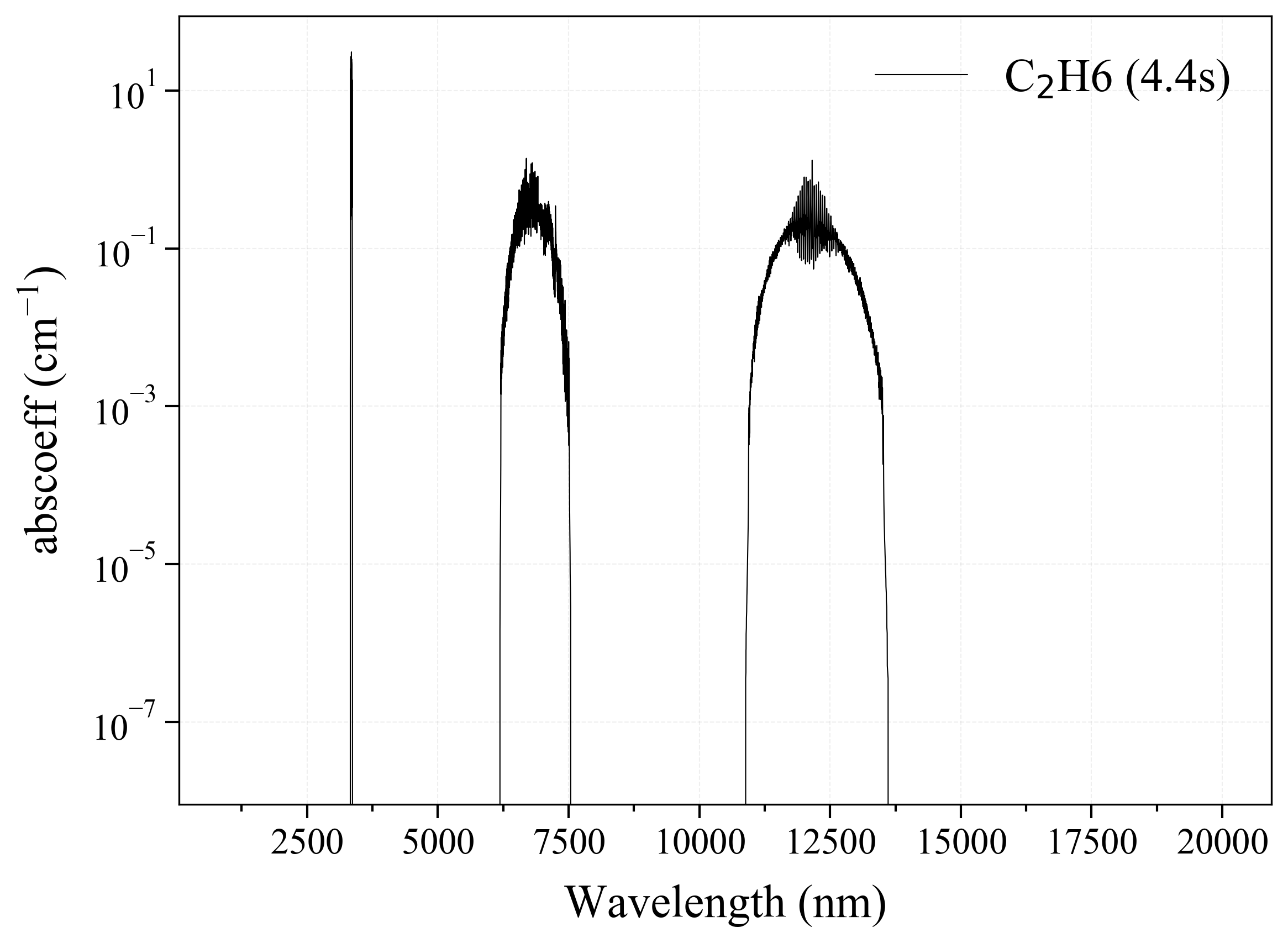
27. C2H6¶
27
'C2H6': Ethane absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='C2H6', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
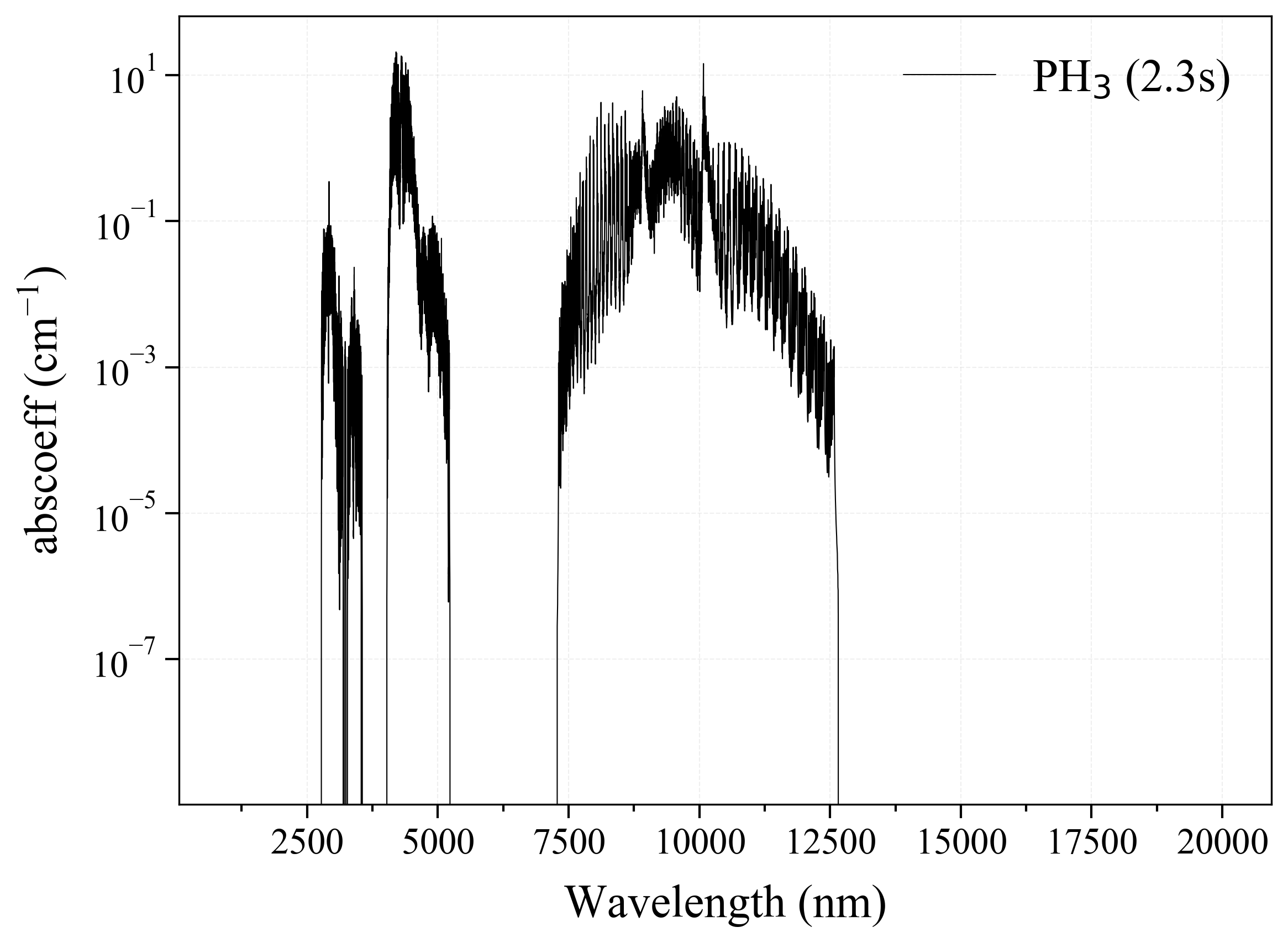
28. PH3¶
28
'PH3': Phosphine absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='PH3', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
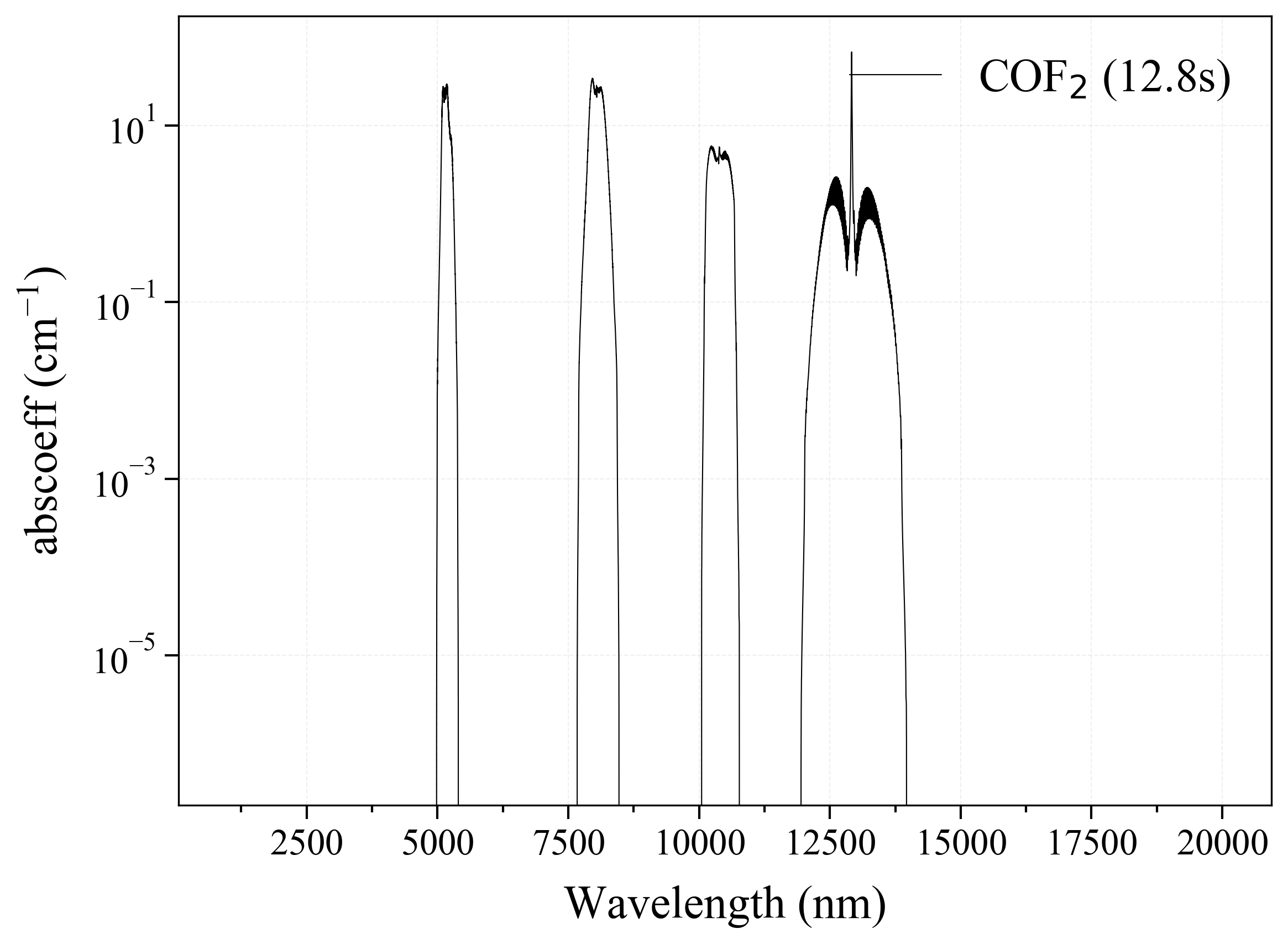
29. COF2¶
29
'COF2': Carbonyl Fluoride absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='COF2', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
30. SF6¶
- 30
'SF6'Sulfur Hexafluoride absorption coefficient (opacity) at 300 Knot calculated.
- 30
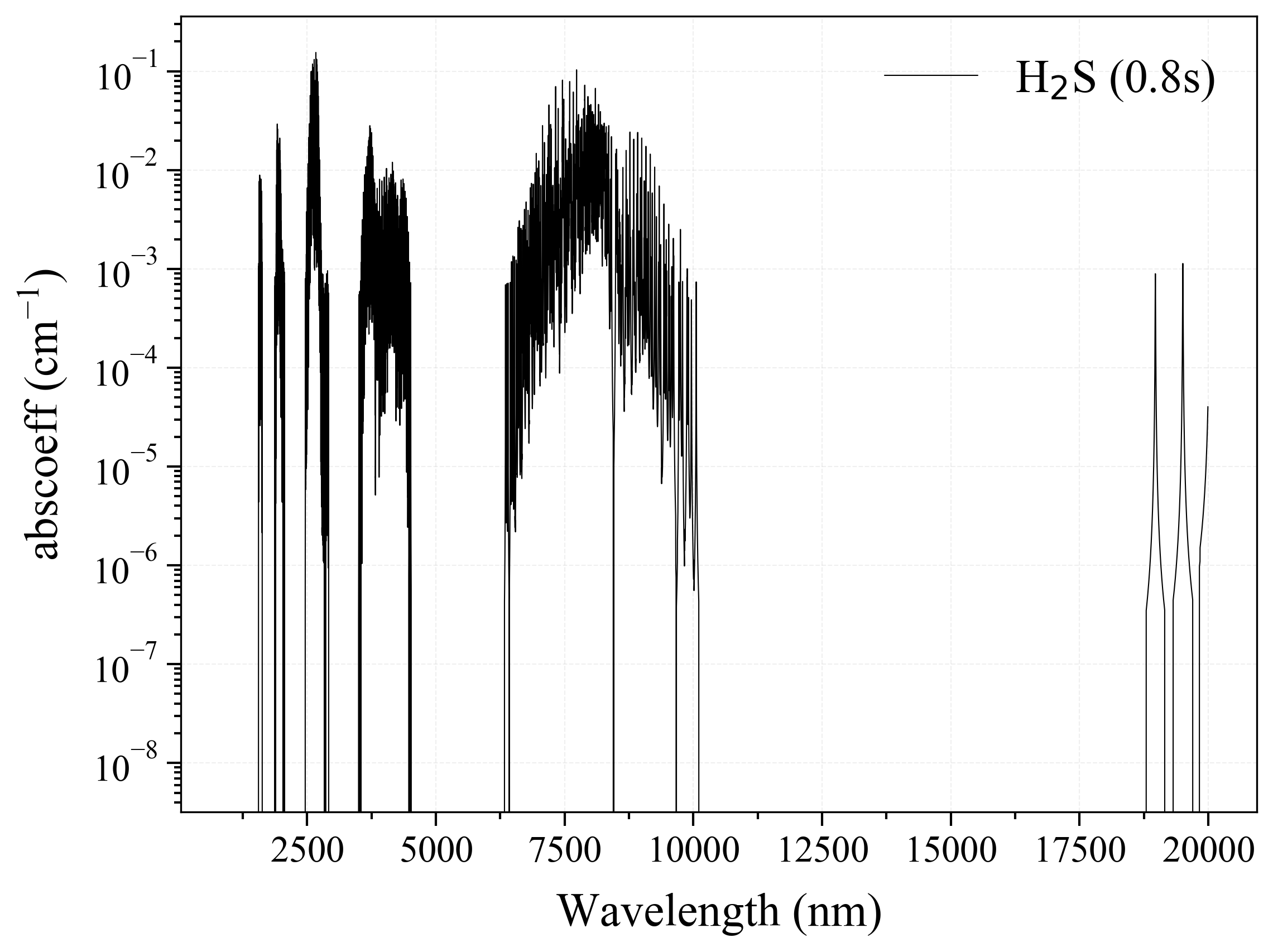
31. H2S¶
31
'H2S': Hydrogen Sulfide absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='H2S', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
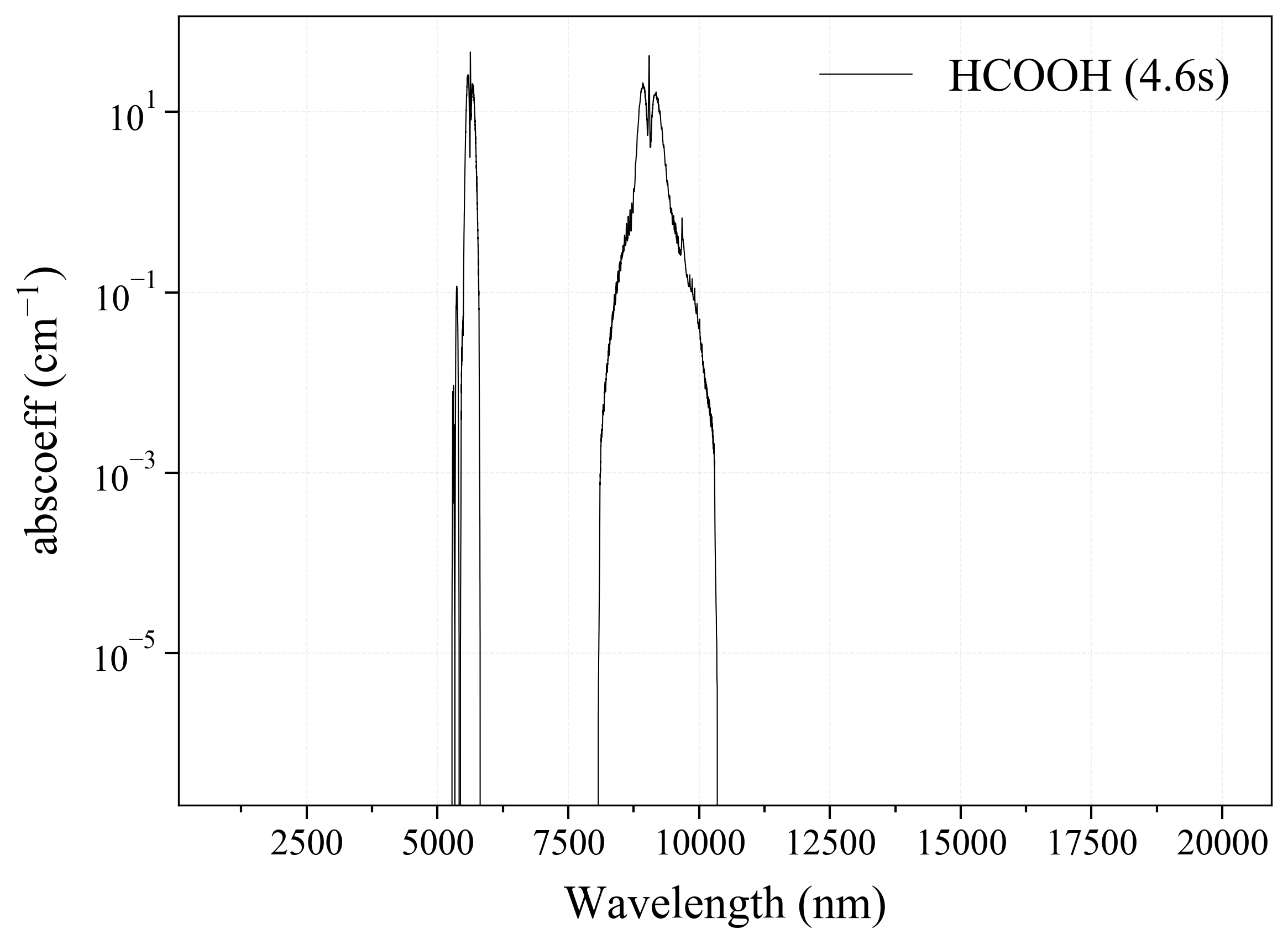
32. HCOOH¶
32
'HCOOH': Formic Acid absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HCOOH', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
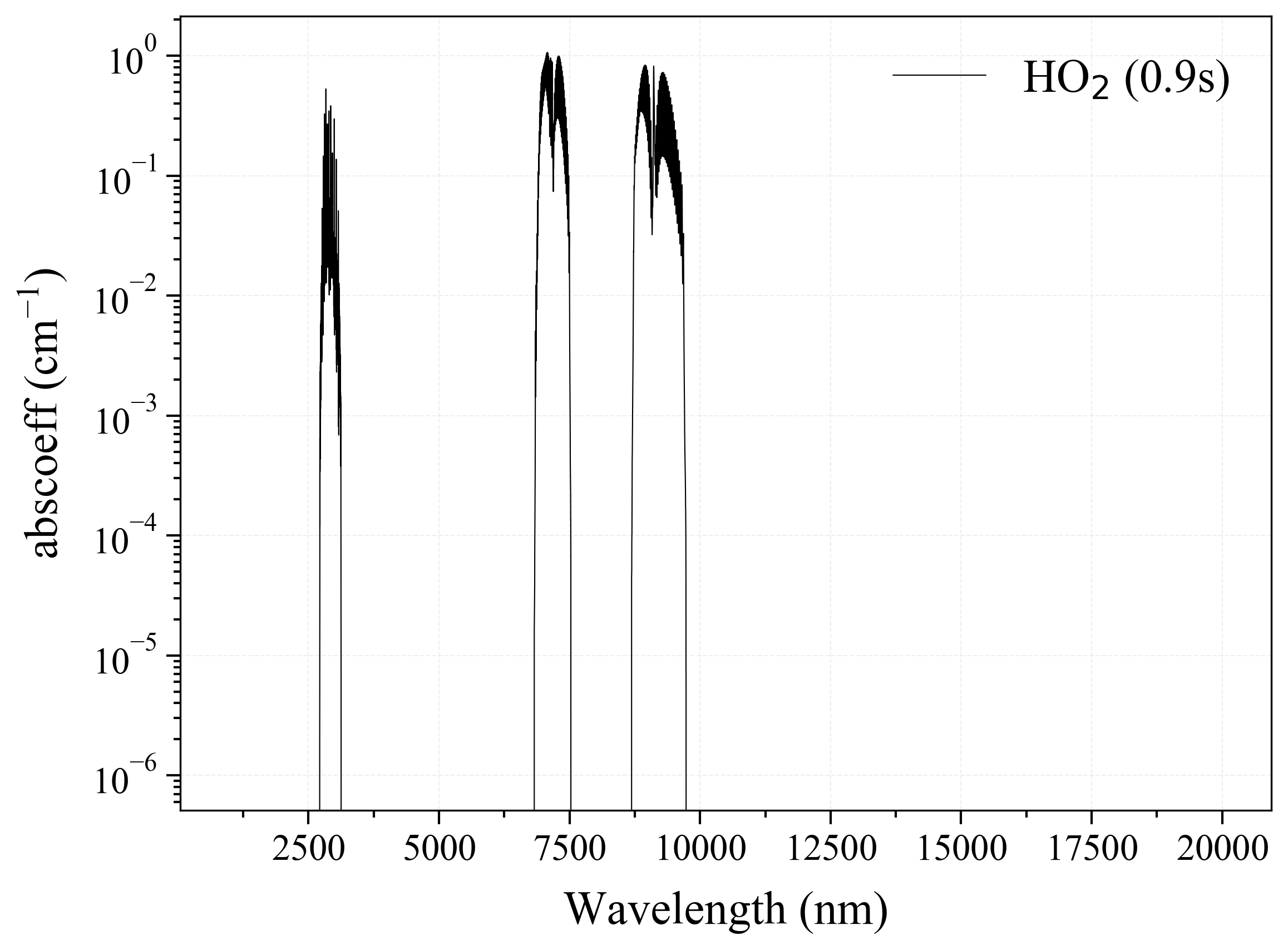
33. HO2¶
33
'HO2': Hydroperoxyl absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='HO2', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
34. O =====-
- 34
'O'Oxygen Atom absorption coefficient (opacity) at 300 Knot calculated.
- 34
35. ClONO2¶
- 35
'ClONO2'Chlorine Nitrate absorption coefficient (opacity) at 300 Knot calculated.
- 35
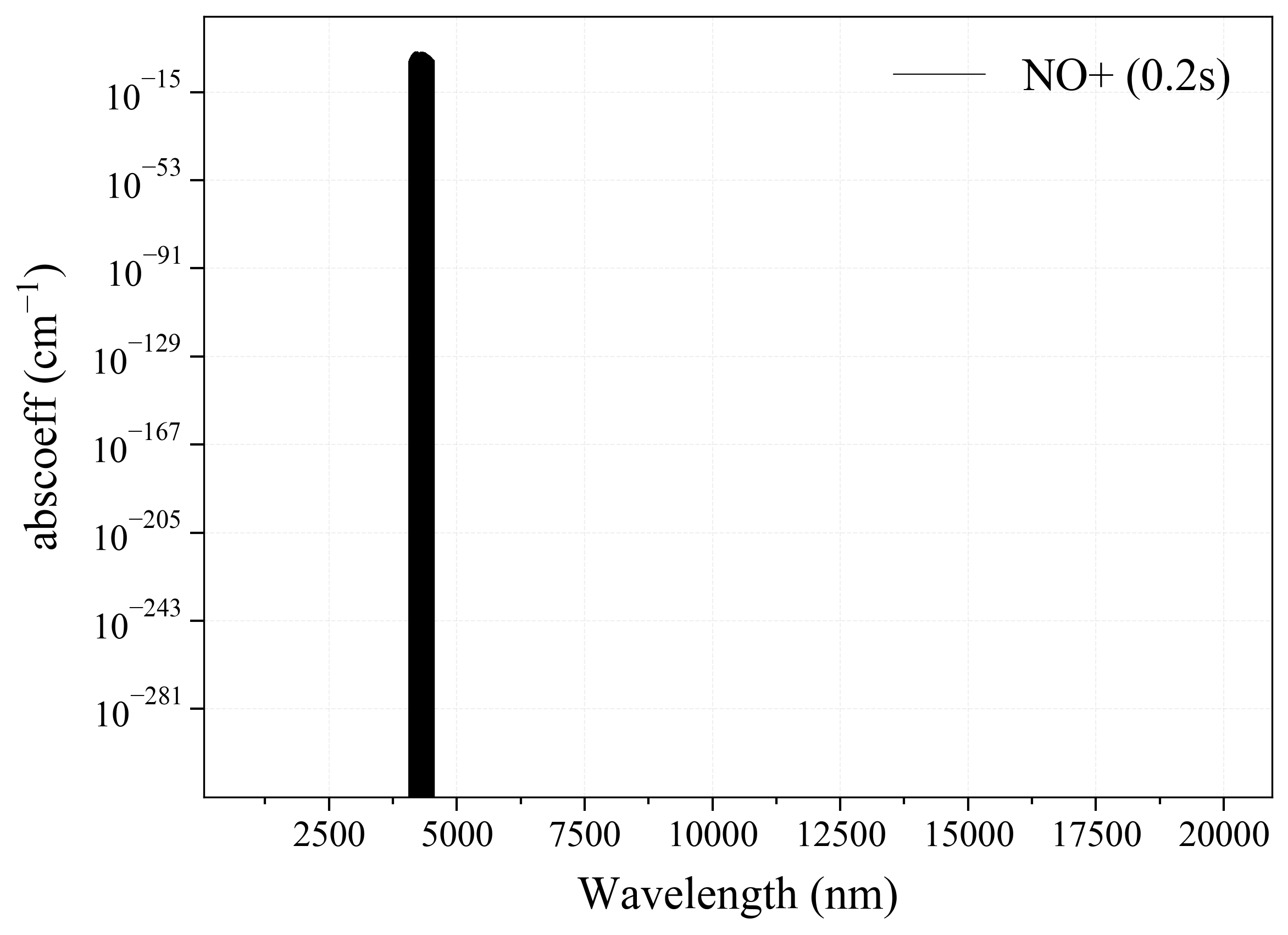
36. NO+¶
36
'NO+': Nitric Oxide Cation absorption coefficient (opacity) at 300 Ks = calc_spectrum(wavelength_min=1000, wavelength_max=20000, Tgas=300, pressure=1, molecule='NO+', optimization=None, cutoff=1e-23, isotope='1') s.plot('abscoeff', wunit='nm')
37. HOBr¶
- 37
'HOBr'Hypobromous Acid absorption coefficient (opacity) at 300 Knot calculated.
- 37
38. C2H4¶
- 38
'C2H4'Ethylene absorption coefficient (opacity) at 300 Knot calculated.
- 38
39. CH3OH¶
- 39
'CH3OH'Methanol absorption coefficient (opacity) at 300 Knot calculated.
- 39
40. CH3Br¶
- 40
'CH3Br'Methyl Bromide absorption coefficient (opacity) at 300 Knot calculated.
- 40
41. CH3CN¶
- 41
'CH3CN'Acetonitrile absorption coefficient (opacity) at 300 Knot calculated.
- 41
42. CF4¶
- 42
'CF4'CFC-14 absorption coefficient (opacity) at 300 Knot calculated.
- 42
43. C4H2¶
- 43
'C4H2'Diacetylene absorption coefficient (opacity) at 300 Knot calculated.
- 43
44. HC3N¶
- 44
'HC3N'Cyanoacetylene absorption coefficient (opacity) at 300 Knot calculated.
- 44
45. H2¶
- 45
'H2'Hydrogen absorption coefficient (opacity) at 300 Knot calculated.
- 45
46. CS¶
- 46
'CS'Carbon Monosulfide absorption coefficient (opacity) at 300 Knot calculated.
- 46
47. SO3¶
- 47
'SO3'Sulfur trioxide absorption coefficient (opacity) at 300 Knot calculated.
- 47
48. C2N2¶
- 48
'C2N2'Cyanogen absorption coefficient (opacity) at 300 Knot calculated.
- 48
49. COCl2¶
- 49
'COCl2'Phosgene absorption coefficient (opacity) at 300 Knot calculated.
- 49



