radis.tools.database module¶
Implements a spectrum database SpecDatabase
class to manage them all.
It basically manages a list of Spectrum JSON files, adding a Pandas dataframe structure on top to serve as an efficient index to visualize the spectra input conditions, and slice through the Dataframe with easy queries
Examples
See and get objects from database:
from radis.tools import SpecDatabase
db = SpecDatabase(r"path/to/database") # create or loads database
db.update() # in case something changed (like a file was added manually)
db.see(['Tvib', 'Trot']) # nice print in console
s = db.get('Tvib==3000 & Trot==1500')[0] # get all spectra that fit conditions
db.add(s) # update database (and raise error because duplicate!)
Note that SpectrumFactory can be configured to
automatically look-up and update a database when spectra are calculated.
An example of script to update all spectra conditions in a database (ex: when a condition was added afterwards to the Spectrum class):
# Example: add the 'medium' key in conditions
db = "database_CO"
for f in os.listdir(db):
if not f.endswith('.spec'): continue
s = load_spec(join(db, f))
s.conditions['medium'] = 'vacuum'
s.store(join(db,f), if_exists_then='replace')
You can see more examples on the Spectrum Database section of the website.
- class SpecDatabase(path='.', filt='.spec', add_info=None, add_date='%Y%m%d', verbose=True, binary=True, nJobs=-2, batch_size='auto', lazy_loading=True, update_register_only=False)[source]¶
Bases:
SpecListA Spectrum Database class to manage them all.
It basically manages a list of Spectrum JSON files, adding a Pandas dataframe structure on top to serve as an efficient index to visualize the spectra input conditions, and slice through the Dataframe with easy queries
Similar to
SpecList, but associated and synchronized with a folder- Parameters:
path (str) – a folder to initialize the database
filt (str) – only consider files ending with
filt. Default.specbinary (boolean) – if
True, open Spectrum files as binary files. IfFalseand it fails, try as binary file anyway. DefaultFalse.lazy_loading (bool``) – If
True, load only the data from the summary csv file and the spectra will be loaded when accessed by the get functions. IfFalse, load all the spectrum files. IfTrueand the summary .csv file does not exist, load all spectra
- Other Parameters:
*input for :class:`~joblib.parallel.Parallel` loading of database*
nJobs (int) – Number of processors to use to load a database (useful for big databases). BE CAREFULL, no check is done on processor use prior to the execution ! Default
-2: use all but 1 processors. Use1for single processor.batch_size (int or
'auto') – The number of atomic tasks to dispatch at once to each worker. When individual evaluations are very fast, dispatching calls to workers can be slower than sequential computation because of the overhead. Batching fast computations together can mitigate this. Default:'auto'More information in :class:`joblib.parallel.Parallel`
Examples
>>> db = SpecDatabase(r"path/to/database") # create or loads database >>> db.update() # in case something changed >>> db.see(['Tvib', 'Trot']) # nice print in console >>> s = db.get('Tvib==3000')[0] # get a Spectrum back >>> db.add(s) # update database (and raise error because duplicate!)
Note that
SpectrumFactorycan be configured to automatically look-up and update a database when spectra are calculated.The function to auto retrieve a Spectrum from database on calculation time is a method of DatabankLoader class
You can see more examples on the Spectrum Database section of the website.
See also
load_spec(),store(),Methods,get(),get_closest(),get_unique(),Methods,see(),update(),add(),compress_to(),find_duplicates(),Compare,fit_spectrum()- add(spectrum: Spectrum, store_name=None, if_exists_then='increment', **kwargs)[source]¶
Add Spectrum to database, whether it’s a
Spectrumobject or a file that stores one. Check it’s not in database already.- Parameters:
spectrum (
Spectrumobject, or path to a .spec file (str)) – if aSpectrumobject: stores it in the database (using thestore()method), then adds the file to the database folder. if a path to a file (str): first copy the file to the database folder, then loads the copied file to the database.- Other Parameters:
store_name (
str, orNone) – name of the file where the spectrum will be stored. IfNone, name is generated automatically from the Spectrum conditions (seeadd_info=andif_exists_then=)if_exists_then (
'increment','replace','error','ignore') – what to do if file already exists. If'increment'an incremental digit is added. If'replace'file is replaced (!). If'ignore'the Spectrum is not added to the database and no file is created. If'error'(or anything else) an error is raised. Default'increment'.**kwargs (**dict) – extra parameters used in the case where spectrum is a file and a .spec object has to be created (useless if
spectrumis a file already). kwargs are forwarded to Spectrum.store() method. See thestore()method for more information.Note
Other
store()parameters can be given as kwargs arguments. See below :compress (0, 1, 2) – if
Trueor 1, save the spectrum in a compressed formif 2, removes all quantities that can be regenerated with
update(), e.g, transmittance if abscoeff and path length are given, radiance if emisscoeff and abscoeff are given in non-optically thin case, etc. If not given, use the value ofSpecDatabase.binaryThe performances are usually better if compress = 2. See https://github.com/radis/radis/issues/84.add_info (list) – append these parameters and their values if they are in conditions example:
nameafter = ['Tvib', 'Trot']
discard (list of str) – parameters to exclude. To save some memory for instance Default
['lines', 'populations']: retrieved Spectrum will loose theline_survey()andplot_populations()methods (but it saves a ton of memory!).
Examples
from radis.tools import SpecDatabase db = SpecDatabase(r"path/to/database") # create or loads database db.add(s, discard=['populations'])
You can see more examples on the Spectrum Database section of the website.
See also
- compress_to(new_folder, compress=True, if_exists_then='error')[source]¶
Saves the Database in a new folder with all Spectrum objects under compressed (binary) format. Read/write is much faster. After the operation, a new database should be initialized in the new_folder to access the new Spectrum.
- Parameters:
new_folder (str) – folder where to store the compressed SpecDatabase. If doesn’t exist, it is created.
compress (boolean, or 2) – if
True, saves under binary format. Faster and takes less space. If2, additionally remove all redundant quantities.if_exists_then (
'increment','replace','error','ignore') – what to do if file already exists. If'increment'an incremental digit is added. If'replace'file is replaced (!). If'ignore'the Spectrum is not added to the database and no file is created. If'error'(or anything else) an error is raised. Default'error'.
See also
- find_duplicates(columns=None)[source]¶
Find spectra with same conditions. The first duplicated spectrum will be
'False', the following will be'True'(see .duplicated()).- Parameters:
columns (list, or
None) – columns to find duplicates on. IfNone, use all conditions.
Examples
db.find_duplicates(columns={'x_e', 'x_N_II'}) Out[34]: file 20180710_101.spec True 20180710_103.spec True dtype: bool
You can see more examples in the Spectrum Database section
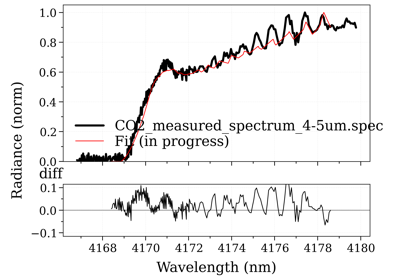
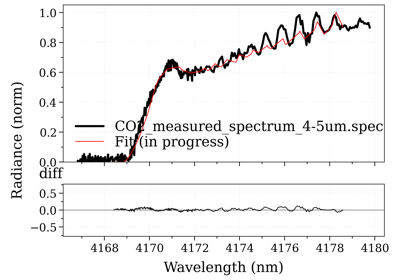
- fit_spectrum(s_exp, residual=None, normalize=False, normalize_how='max', conditions='', **kwconditions)[source]¶
Returns the Spectrum in the database that has the lowest residual with
s_exp.- Parameters:
s_exp (Spectrum) –
Spectrumto fit (typically: experimental spectrum)- Other Parameters:
residual (func, or
None) – which residual function to use. IfNone, useget_residual()with optionignore_nan=Trueand optionsnormalizeandnormalize_howas defined by the user.get_residualshould have the form:lambda s_exp, s, normalize: func(s_exp, s, normalize=normalize)
where the output is a float. Default
Noneconditions, **kwconditions (str, **dict) – restrain fitting to only Spectrum that match the given conditions in the database. See
get()for more information.normalize (bool, or Tuple) – see
get_residual()normalize_how (‘max’, ‘area’) – see
get_residual()
- Returns:
s_best – closest Spectrum to
s_exp- Return type:
Examples
Using a customized residual function (below: to get the transmittance):
from radis import get_residual db = SpecDatabase('...') db.fit_spectrum(s_exp, get_residual=lambda s_exp, s: get_residual(s_exp, s, var='transmittance'))
You can see more examples on the Spectrum Database section More advanced tools for interactive fitting of multi-dimensional, multi-slabs spectra can be found in
fitroom.See also
- interpolate(**kwconditions)[source]¶
Interpolate existing spectra from the database to generate a new spectrum with conditions kwargs
Examples
db.interpolate(Tgas=300, mole_fraction=0.3)
- to_dict()[source]¶
Returns all Spectra in database under a dictionary, indexed by file.
- Returns:
out – {path : Spectrum object} dictionary
- Return type:
dict
Note
SpecList.items().values()is equivalent toSpecList.get()
- update(force_reload=False, filt='.spec', update_register_only=False)[source]¶
Reloads database, updates internal index structure and export it in
<database>.csv.- Parameters:
force_reload (boolean) – if
True, reloads files already in database. DefaultFalsefilt (str) – only consider files ending with
filt. Default.spec
- Other Parameters:
update_register_only (bool) – if
True, load files and update csv but do not keep the Spectrum in memory. DefaultFalse
Notes
Can be loaded in parallel using joblib by setting the
nJobsandbatch_sizeattributes ofSpecDatabase. Seejoblib.parallel.Parallelfor information on the arguments
- class SpecList(*spectra, **kwargs)[source]¶
Bases:
object- create_fname_grid(conditions)[source]¶
Create a 2D-grid of filenames for the list of parameters
conditionsExamples
db.create_fname_grid(["Tgas", "pressure"])
See also
- get(conditions='', **kwconditions)[source]¶
Returns a list of spectra that match given conditions.
- Parameters:
database (list of Spectrum objects) – the database
conditions (str) –
a list of conditions. Example:
db.get('Tvib==3000 & Trot==1500')
kwconditions (dict) –
an unfolded dict of conditions. Example:
db.get(Tvib=3000, Trot=1500)
- Other Parameters:
inplace (
bool) – if True, return the actual object in the database. Else, return copies. DefaultFalseverbose (
bool) – more blablascale_if_possible (
bool) – ifTrue, spectrum is scaled for parameters that can be computed directly from spectroscopic quantities (e.g:'path_length','molar_fraction'). DefaultFalse
- Returns:
out
- Return type:
list of Spectrum
Examples
spec_list = db.get('Tvib==3000 & Trot==1300')
or:
spec_list = db.get(Tvib=3000, Trot=1300)
See also
get_unique(),get_closest(), ;py:meth:interpolate,items()
- get_closest(scale_if_possible=True, **kwconditions)[source]¶
Returns the Spectra in the database that is the closest to the input conditions.
Note that for non-numeric values only equals should be given. To calculate the distance all numeric values are scaled by their mean value in the database
- Parameters:
kwconditions (named arguments) – i.e:
Tgas=300, path_length=1.5scale_if_possible (boolean) – if
True, spectrum is scaled for parameters that can be computed directly from spectroscopic quantities (e.g:'path_length','molar_fraction'). DefaultTrue
- Other Parameters:
verbose (boolean) – print messages. Default
Trueinplace (boolean) – if
True, returns the actual object in database. Else, return a copy. DefaultFalse
See also
get(),get_unique(), ;py:meth:interpolate
- get_items(condition)[source]¶
Returns all Spectra in database under a dictionary; indexed by
conditionRequires that
conditionis unique- Parameters:
condition (str) – condition. Ex:
Trot- Returns:
out – {condition:Spectrum}
- Return type:
dict
Examples
db.get_items("Tgas")
See also
- get_unique(conditions='', scale_if_possible=False, **kwconditions)[source]¶
Returns a spectrum that match given conditions.
Raises an error if the spectrum is not unique.
See also
get(),get_closest(), ;py:meth:interpolate
- items()[source]¶
Iterate over all
Spectrumin database.Examples
Print name of all Spectra in dictionary:
db = SpecDatabase('.') for path, s in db.items(): print(path, s.name)
Update all spectra in current folder with a new condition (‘author’):
db = SpecDatabase('.') for path, s in db.items(): s.conditions['author'] = 'me' s.store(path, if_exists_then='replace')
- map(function)[source]¶
Apply
functionto all Spectra in database.Examples
Add a missing parameter:
db = SpecDatabase('...') def add_condition(s): s.conditions['exp_run'] = 1 return s db.map(add_condition)
Note
spectra are not changed on disk. If you want to update on disk you may want to combine map() followed by
compress_to()Example
# See length of all spectra : db.map(lambda s: print(len(s))) # Resample all on spectrum of minimum wstep s_wstep_min = db.get(wstep=float(db.see("wstep").min()))[0] db.map(lambda s: s.resample(s_wstep_min)) # Export to a new database: db.compress_to(db.path+'_interp')
- plot(nfig=None, legend=True, **kwargs)[source]¶
Plot all spectra in database.
- Parameters:
nfig (str, or int, or
None) – figure to plot on. DefaultNone: creates one- Other Parameters:
kwargs (dict) – parameters forwarded to the Spectrum
plot()methodlegend (bool) – if
True, plot legend.
- Returns:
fig, ax – figure
- Return type:
matplotlib figure and ax
Examples
Plot all spectra in a folder:
db = SpecDatabase('my_folder') db.plot(wunit='nm')
See also
Spectrum
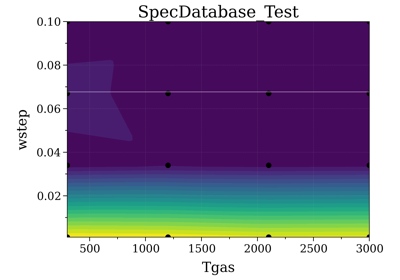
- plot_cond(cond_x, cond_y, z_value=None, nfig=None)[source]¶
Plot database conditions available:
- Parameters:
cond_x, cond_y (str) – columns (conditions) of database.
z_value (array, or None) – if not None, colors the 2D map with z_value. z_value is ordered so that z_value[i] corresponds to row[i] in database.
Examples
- ::
>>> db.plot(Tvib, Trot) # plot all points calculated
>>> db.plot(Tvib, Trot, residual) # where residual is calculated by a fitting # procedure...
- see(columns=None, *args)[source]¶
Shows Spectrum database with all conditions (
columns=None) or specific conditions.- Parameters:
columns (str, list of str, or None) – shows the conditions value for all cases in database. If None, all conditions are shown. Default
Nonee.g.:db.see(['Tvib', 'Trot'])
Notes
Makes the ‘file’ column the index, and also discard the ‘Spectrum’ column (that holds all the data) for readability
- load_spec(file, binary=True) Spectrum[source]¶
Loads a .spec file into a
Spectrumobject. Addsfilein the Spectrumfileattribute.- Parameters:
file (str) – .spec file to load
binary (boolean) – set to
Trueif the file is encoded as binary. DefaultTrue. Will autodetect if it fails, but that may take longer.
- Returns:
Spectrum
- Return type:
a
Spectrumobject
Examples
Example #3: non-equilibrium spectrum (Tvib, Trot, x_CO)
Example #3: non-equilibrium spectrum (Tvib, Trot, x_CO)See also
- plot_spec(file, what='radiance', title=True, **kwargs)[source]¶
Plot a .spec file. Uses the
plot()method internally.- Parameters:
file (str, or Spectrum object) – .spec file to load, or Spectrum object directly
- Other Parameters:
kwargs (dict) – arguments forwarded to
plot()- Returns:
fig – where the Spectrum has been plotted
- Return type:
matplotlib figure
See also
- read_conditions_file(path, verbose=True)[source]¶
Read .csv file with calculation/measurement conditions of all spectra.
File must have at least the column “file”
- Parameters:
path (csv file) – summary of all spectra conditions.
- Return type:
None.
- save(s: Spectrum, path, discard=[], compress=True, add_info=None, add_date=None, if_exists_then='increment', verbose=True, warnings=True)[source]¶
Save a
Spectrumobject in JSON format. Object can be recovered withload_spec(). If manySpectrumare saved in a same folder you can view their properties with theSpecDatabasestructure.- Parameters:
s (Spectrum) – to save
path (str) – filename to save. No extension needed. If filename already exists then a digit is added. If filename is a directory then a new file is created within this directory.
discard (list of str) – parameters to discard. To save some memory.
compress (boolean) – if
False, save under text format, readable with any editor. ifTrue, saves under binary format. Faster and takes less space. If2, removes all quantities that can be regenerated with s.update(), e.g, transmittance if abscoeff and path length are given, radiance if emisscoeff and abscoeff are given in non-optically thin case, etc. DefaultFalseadd_info (list, or None/False) – append these parameters and their values if they are in conditions. e.g:
add_info = ['Tvib', 'Trot']
add_date (str, or
None/False) – adds date in strftime format to the beginning of the filename. e.g:add_date = '%Y%m%d'
if_exists_then (
'increment','replace','error','ignore') – what to do if file already exists. If'increment'an incremental digit is added. If'replace'file is replaced (!). If'ignore'the Spectrum is not added to the database and no file is created. If'error'(or anything else) an error is raised. Default'increment'.
- Returns:
fout – filename used (may be different from given path as new info or incremental identifiers are added)
- Return type:
str
See also