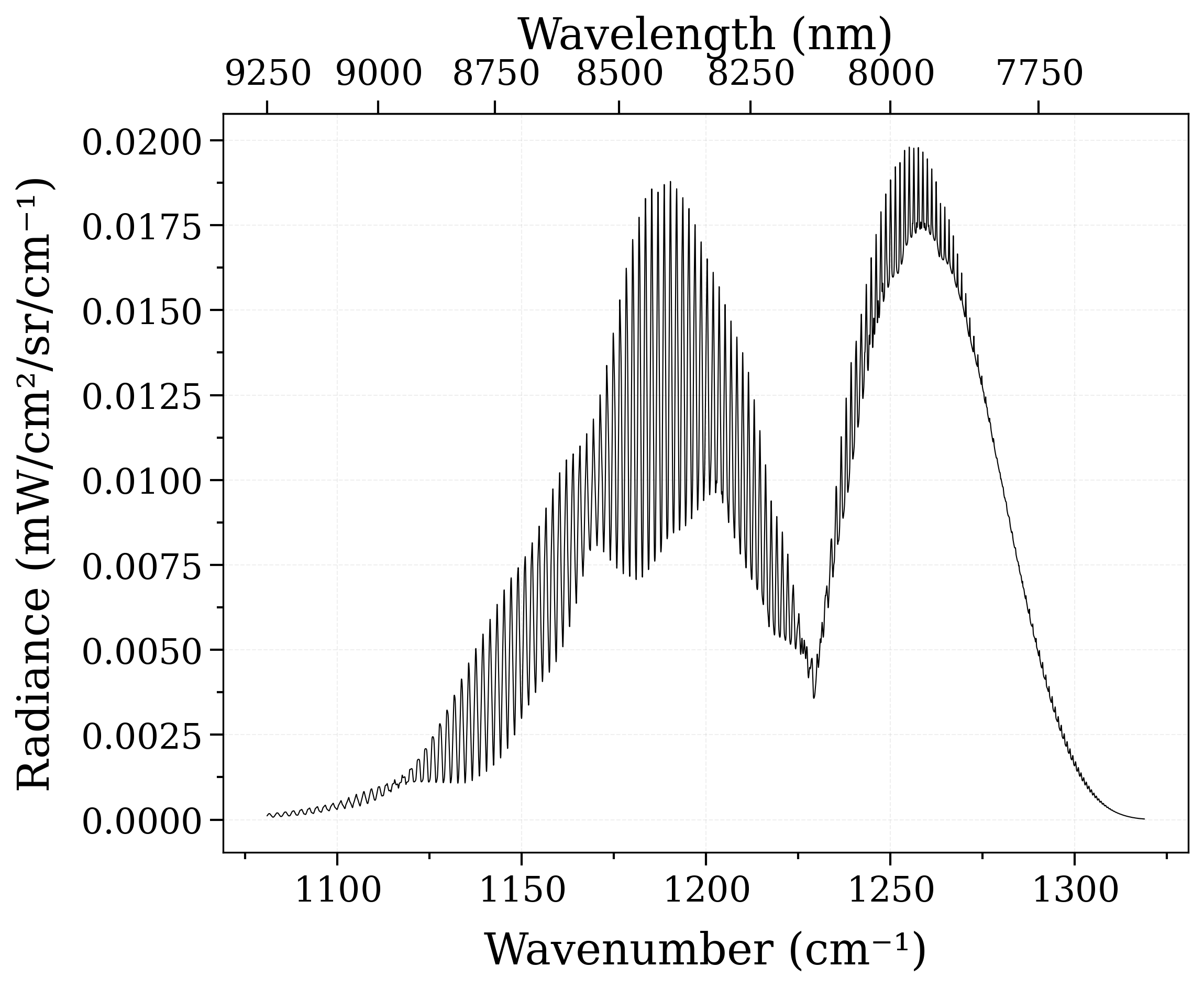
Calculate a spectrum from ExoMol¶
Auto-download and compute a SiO spectrum from the ExoMol database ([ExoMol-2020])
from radis import calc_spectrum
s = calc_spectrum(
1080,
1320, # cm-1
molecule="SiO",
isotope="1",
pressure=1.01325, # bar
Tgas=1000, # K
mole_fraction=0.1,
path_length=1, # cm
databank=("exomol", "EBJT"), # Simply use 'exomol' for the recommended database
)
s.apply_slit(1, "cm-1") # simulate an experimental slit
s.plot("radiance")
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
========== Loading Exomol database [start] ==========
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/EBJT/28Si-16O__EBJT.def
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/EBJT/28Si-16O__EBJT.pf
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/EBJT/28Si-16O__EBJT.states.bz2
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__H2.broad
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__He.broad
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__air.broad
Error: Couldn't download .broad file at http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__air.broad and save.
Note: Caching states data to the vaex format. After the second time, it will become much faster.
Molecule: SiO
Isotopologue: 28Si-16O
Background atmosphere: Air
ExoMol database: EBJT
Local folder: /home/docs/.radisdb/exomol/SiO/28Si-16O/EBJT
Transition files:
=> File 28Si-16O__EBJT.trans
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/EBJT/28Si-16O__EBJT.trans.bz2
=> Caching the *.trans.bz2 file to the vaex (*.h5) format. After the second time, it will become much faster.
=> You can deleted the 'trans.bz2' file by hand.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/exomolapi.py:1258: UserWarning:
Could not load `28Si-16O__Air.broad`. The default broadening parameters are used.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
========== Loading Exomol database [end] ==========
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: MissingPressureShiftWarning:
Pressure-shift coefficient not given in database: assumed 0 pressure shift
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1
mole_fraction 0.1
molecule SiO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 1320.0000 cm-1
wavenum_min 1080.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat exomol-radisdb
dbpath /home/docs/.radisdb/exomol/SiO/28Si-16O/EBJT
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt exomol
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 24001
----------------------------------------
0.03s - Spectrum calculated
<matplotlib.lines.Line2D object at 0x7fbde62d78b0>
"""ExoMol lines can be downloaded and accessed separately using
:py:func:`~radis.io.exomol.fetch_exomol`
"""
# See line data:
from radis.io.exomol import fetch_exomol
df = fetch_exomol("SiO", database="EBJT", isotope="1", load_wavenum_max=5000)
print(df)
========== Loading Exomol database [start] ==========
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__H2.broad
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__He.broad
=> Downloading from http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__air.broad
Error: Couldn't download .broad file at http://www.exomol.com/db/SiO/28Si-16O/28Si-16O__air.broad and save.
Molecule: SiO
Isotopologue: 28Si-16O
Background atmosphere: Air
ExoMol database: EBJT
Local folder: /home/docs/.radisdb/exomol/SiO/28Si-16O/EBJT
Transition files:
=> File 28Si-16O__EBJT.trans
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/exomolapi.py:1258: UserWarning:
Could not load `28Si-16O__Air.broad`. The default broadening parameters are used.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
========== Loading Exomol database [end] ==========
i_upper i_lower A ... int airbrd Tdpair
0 24300 24299 1.926600e-12 ... 3.923990e-167 0.07 0.5
1 4698 5073 3.431600e-10 ... 2.626438e-202 0.07 0.5
2 24273 24272 1.652000e-10 ... 2.386983e-165 0.07 0.5
3 24301 24300 1.725200e-11 ... 2.947629e-166 0.07 0.5
4 24232 24231 7.675700e-10 ... 1.291448e-164 0.07 0.5
... ... ... ... ... ... ... ...
249599 3769 1818 2.429700e-04 ... 3.947150e-91 0.07 0.5
249600 8052 6269 1.481800e-02 ... 2.484685e-72 0.07 0.5
249601 8693 6938 1.601100e-02 ... 1.337757e-67 0.07 0.5
249602 8659 6904 1.166600e-02 ... 3.267363e-66 0.07 0.5
249603 7665 5871 3.160000e-03 ... 4.399687e-64 0.07 0.5
[249604 rows x 11 columns]
See the list of recommended databases for the 1st isotope of SiO :
from radis.io.exomol import get_exomol_database_list, get_exomol_full_isotope_name
databases, recommended = get_exomol_database_list(
"SiO", get_exomol_full_isotope_name("SiO", 1)
)
print("Databases for SiO: ", databases)
print("Database recommended by ExoMol: ", recommended)
Databases for SiO: ['Kurucz-SiO', 'EBJT', 'xsec-SiOUVenIR', 'SiOUVenIR']
Database recommended by ExoMol: SiOUVenIR
Total running time of the script: (0 minutes 9.557 seconds)



