GPU Accelerated Spectra¶
Example using GPU calculation with eq_spectrum_gpu()
This method requires a GPU - Currently, only Nvidia GPU’s are supported. For more information on how to setup your system to run GPU-accelerated methods using CUDA, check GPU Spectrum Calculation on RADIS
Note
in the example below, the code runs on the GPU by default. In case no Nvidia GPU is
detected, the code will instead be ran on CPU. This can be toggled manually by setting
the backend keyword either to 'gpu-cuda' or 'cpu-cuda'.
The run time reported below is for CPU.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
In ``/home/docs/.radisdb/hitemp`` keep only relevant input files:
/home/docs/.radisdb/hitemp/CO2-02_02125-02250_HITEMP2010.hdf5
/home/docs/.radisdb/hitemp/CO2-02_02250-02500_HITEMP2010.hdf5
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1500.0 K
Trot 1500.0 K
Tvib 1500.0 K
isotope 1,2,3
mole_fraction 0.8
molecule CO2
overpopulation None
path_length 0.2 cm
pressure 1.0 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2450.0000 cm-1
wavenum_min 2150.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 0 cm-1/(#.cm-2)
dbformat hitemp-radisdb
dbpath /home/docs/.radisdb/hitemp/CO2-02_02125-02250_HITEMP2010.hdf5,/home/docs/.radisdb/hitemp/CO2-02_0225...
diluent air
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm False
truncation 50 cm-1
waveunit cm-1
wstep 0.002 cm-1
zero_padding 150001
----------------------------------------
0.91s - Spectrum calculated
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/lbl/factory.py:1107: FutureWarning:
Calling float on a single element Series is deprecated and will raise a TypeError in the future. Use float(ser.iloc[0]) instead
Can't find libcuda.so.1...
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/gpu/gpu.py:112: NoGPUWarning:
Failed to load CUDA context, this happened either becauseCUDA is not installed properly, or you have no NVIDIA GPU. Continuing with emulated GPU on CPU...This means *NO* GPU acceleration!
Number of lines loaded: 1128265
mode: CPU
Finished calculating spectrum!
0.55s - Spectrum calculated
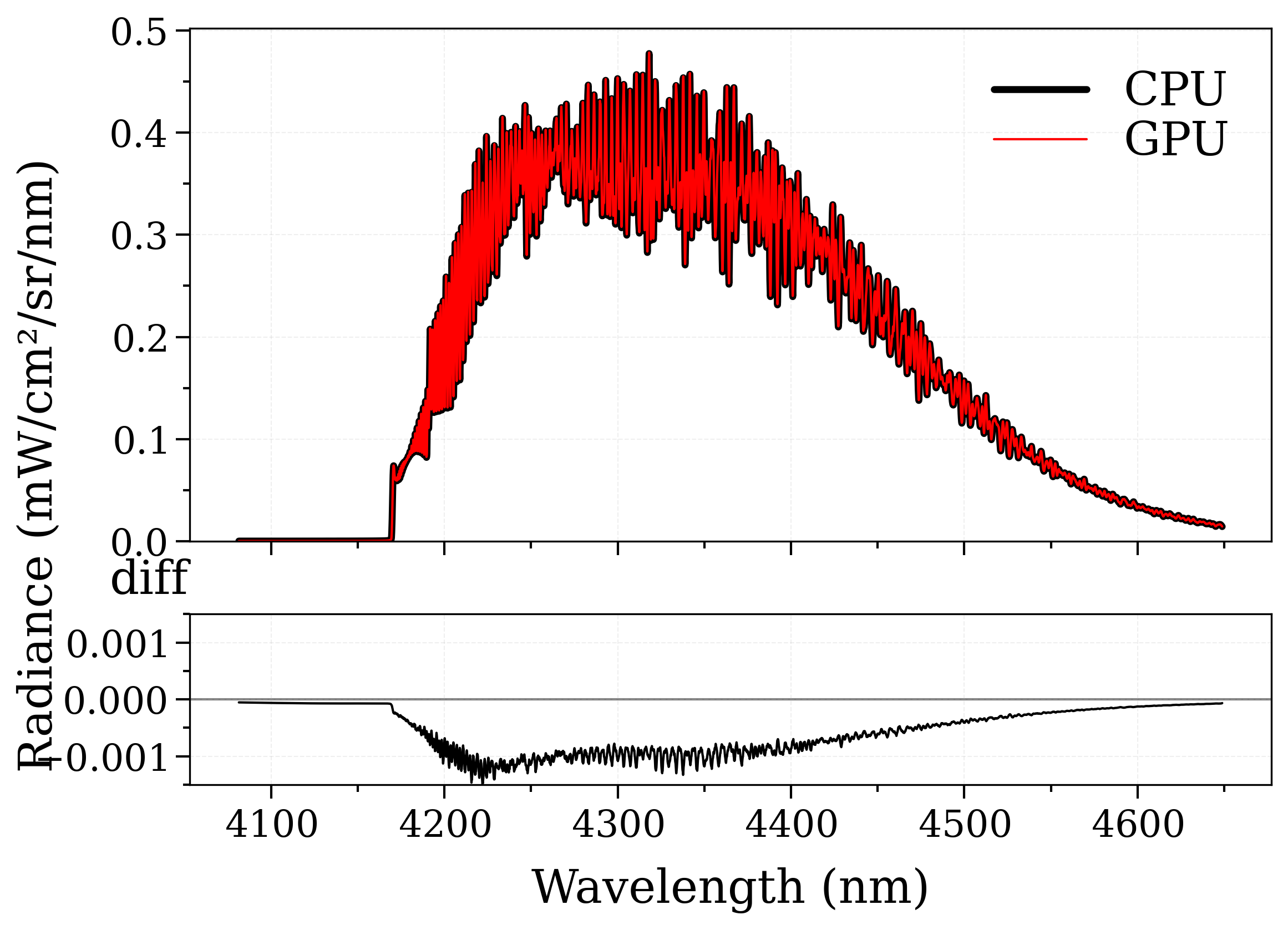
(<Figure size 640x480 with 2 Axes>, [<Axes: >, <Axes: xlabel='Wavelength (nm)'>])
from radis import SpectrumFactory, plot_diff
sf = SpectrumFactory(
2150,
2450, # cm-1
molecule="CO2",
isotope="1,2,3",
wstep=0.002,
)
sf.fetch_databank("hitemp")
T = 1500.0 # K
p = 1.0 # bar
x = 0.8
l = 0.2 # cm
w_slit = 0.5 # cm-1
s_cpu = sf.eq_spectrum(
name="CPU",
Tgas=T,
pressure=p,
mole_fraction=x,
path_length=l,
)
s_cpu.apply_slit(w_slit, unit="cm-1")
s_gpu = sf.eq_spectrum_gpu(
name="GPU",
Tgas=T,
pressure=p,
mole_fraction=x,
path_length=l,
backend="gpu-cuda",
)
s_gpu.apply_slit(w_slit, unit="cm-1")
plot_diff(s_cpu, s_gpu, var="radiance", wunit="nm", method="diff")
Total running time of the script: (0 minutes 3.800 seconds)



