Note
Run this example online :
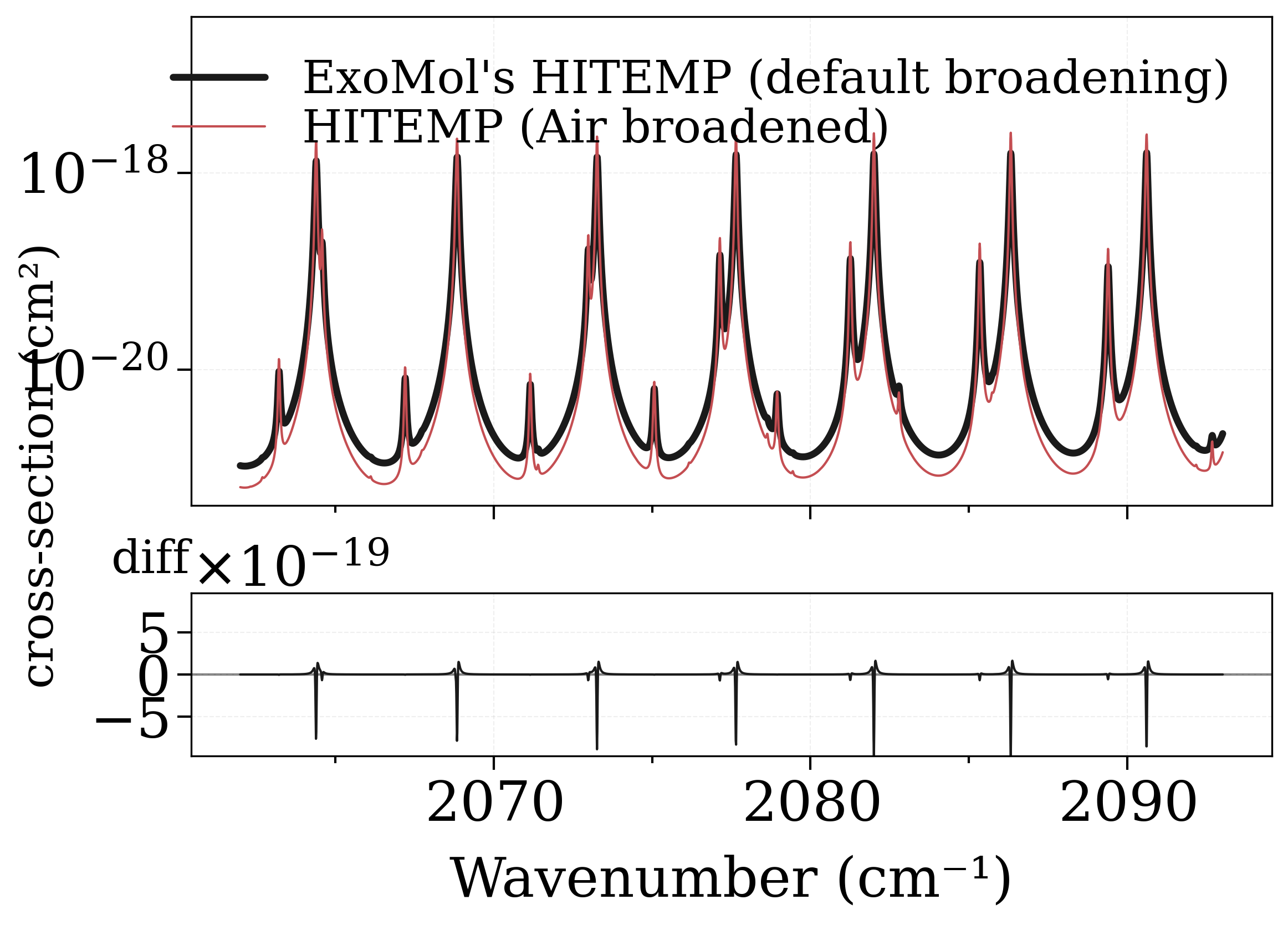
Compare CO xsections from the ExoMol and HITEMP database¶
Auto-download and calculate CO spectrum from the HITEMP database, and the
ExoMol database. ExoMol references multiple databases for CO. Here we do not
use the ExoMol recommended database (see get_exomol_database_list())
but we use the HITEMP database hosted on ExoMol.
Output should be similar, but no ! By default these two databases provide different broadening coefficients. However, the Einstein coefficients & linestrengths should be the same, therefore the integrals under the lines should be similar.
We verifiy this below :
For further exploration, ExoMol and HITEMP lines can be downloaded and accessed separately using
fetch_exomol() and fetch_hitemp()
import astropy.units as u
from radis import calc_spectrum, plot_diff
conditions = {
"wmin": 2062 / u.cm,
"wmax": 2093 / u.cm,
"molecule": "CO",
"isotope": "1",
"pressure": 1.01325, # bar
"Tgas": 1000, # K
"mole_fraction": 0.1,
"path_length": 1, # cm
"broadening_method": "fft", # @ dev: Doesn't work with 'voigt'
"verbose": True,
"neighbour_lines": 20,
}
Note that we account for the effect on neighbour_lines by computing 20cm-1
on the side (neighbour_lines condition above)
s_exomol = calc_spectrum(
**conditions, databank="exomol", name="ExoMol's HITEMP (default broadening)"
)
s_hitemp = calc_spectrum(
**conditions,
databank="hitemp",
name="HITEMP (Air broadened)",
)
fig, [ax0, ax1] = plot_diff(s_exomol, s_hitemp, "xsection", yscale="log")
# Adjust diff plot to be in linear scale
ax1.set_yscale("linear")
ax0.set_ylim(ymax=ax0.get_ylim()[1] * 10) # more space for legend
Using ExoMol database Li2015 for 12C-16O (recommended by the ExoMol team). All available databases are ['HITEMP', 'Li', 'xsec-Li2015', 'xsec-VUV-DTU', 'Li2015']. Select one of them with `radis.fetch_exomol(DATABASE_NAME)`, `SpectrumFactory.fetch_databank('exomol', exomol_database=DATABASE_NAME')`, or `calc_spectrum(..., databank=('exomol', DATABASE_NAME))`
Background atmosphere: Air
Downloading http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.def
Downloading http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.pf
Downloading http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.states.bz2
Downloading http://www.exomol.com/db/CO/12C-16O/12C-16O__H2.broad
Downloading http://www.exomol.com/db/CO/12C-16O/12C-16O__He.broad
Downloading http://www.exomol.com/db/CO/12C-16O/12C-16O__air.broad
Error: Couldn't download .broad file at http://www.exomol.com/db/CO/12C-16O/12C-16O__air.broad and save.
Note: Caching states data to the vaex format. After the second time, it will become much faster.
Reading transition file
Downloading http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.trans.bz2
Note: Caching line transition data to the vaex format. After the second time, it will become much faster.
.broad is used.
Warning: Cannot load .broad. The default broadening parameters are used.
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2093.0000 cm-1
wavenum_min 2062.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used
broadening_method fft
cutoff 1e-27 cm-1/(#.cm-2)
dbformat exomol-radisdb
dbpath /home/docs/.radisdb/exomol/CO/12C-16O/Li2015
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 20 cm-1
optimization simple
parfuncfmt exomol
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm auto
truncation None cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding -1
----------------------------------------
/home/docs/checkouts/readthedocs.org/user_builds/radis/envs/master/lib/python3.8/site-packages/radis/misc/warning.py:354: MissingPressureShiftWarning:
Pressure-shift coefficient not given in database: assumed 0 pressure shift
0.02s - Spectrum calculated
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure_mbar 1013.25 mbar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2093.0000 cm-1
wavenum_min 2062.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used
broadening_method fft
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitemp-radisdb
dbpath /home/docs/.radisdb/hitemp/CO-05_HITEMP2019.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 20 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm auto
truncation None cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding -1
----------------------------------------
0.02s - Spectrum calculated
(4.1789321827835834e-22, 3.8605771326502384e-17)
Broadening coefficients are different in these databases, so lineshapes end up being very different; however the areas under the lines should be the same. We verify this :
import numpy as np
try:
assert np.isclose(
s_exomol.get_integral("xsection"), s_hitemp.get_integral("xsection"), rtol=0.001
)
except:
# @dev: someone there is un expected error with this example on ReadThedocs.
# Escaping for the moment. See https://github.com/radis/radis/issues/501
pass
Total running time of the script: ( 0 minutes 7.079 seconds)



