Note
Run this example online :
Calculate a large spectrum by part¶
In may be faster to calculate a full-range spectrum on several partial spectral ranges, and combine them.
Uses the MergeSlabs() function for that.
Starting from 0.11, RADIS introduces a sparse waverange implementation that should make it possible to directly compute a full range spectrum. See the HITRAN full-range example
OH-hitemp-radisdb-1000K-#5120 new quantities added: ['emisscoeff']
OH-hitemp-radisdb-1000K-#5376 new quantities added: ['emisscoeff']
OH-hitemp-radisdb-1000K-#5632 new quantities added: ['emisscoeff']
Spectrum Name: OH-hitemp-radisdb-1000K-#5120//OH-hitemp-radisdb-1000K-#5376//OH-hitemp-radisdb-1000K-#5632
Spectral Quantities
----------------------------------------
abscoeff [cm-1] (99,502 points)
emisscoeff [mW/cm3/sr/cm-1] (99,502 points)
radiance_noslit [mW/cm2/sr/cm-1] (99,502 points)
emissivity_noslit (99,502 points)
absorbance (99,502 points)
transmittance_noslit (99,502 points)
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope N/A
mole_fraction 0.1
molecule {'OH'}
overpopulation None
path_length 1 cm
pressure_mbar 10000.0 mbar
rot_distribution boltzmann
self_absorption True
state X
thermal_equilibrium True
vib_distribution boltzmann
wavenum_max N/A cm-1
wavenum_min N/A cm-1
Computation Parameters
----------------------------------------
NwG N/A
NwL 4
Tref 296 K
add_at_used cython
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitemp-radisdb
dbpath /home/docs/.radisdb/hitemp/OH-13_HITEMP2020.hdf5
default_output_unit cm-1
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
profiler N/A
pseudo_continuum_threshold 0
radis_version 0.13.1
sparse_ldm True
spectral_points N/A
truncation 50 cm-1
waveunit cm-1
wstep 0.1 cm-1
zero_padding N/A
Config parameters
----------------------------------------
DEFAULT_DOWNLOAD_PATH ~/.radisdb
GRIDPOINTS_PER_LINEWIDTH_ERROR_THRESHOLD 1
GRIDPOINTS_PER_LINEWIDTH_WARN_THRESHOLD 3
SPARSE_WAVERANGE auto
Information
----------------------------------------
calculation_time 0.22960855498968158 s
chunksize None
db_use_cached True
dxG 0.13753507880165727
dxL 0.20180288881201608
export_lines False
export_populations None
export_rovib_fraction True
levelsfmt None
lines_calculated 5,672
lines_cutoff 21,775
lines_in_continuum 0
load_energies False
lvl_use_cached True
parfuncpath None
total_lines 27447
warning_broadening_threshold 0.01
warning_linestrength_cutoff 0.01
wavenum_max_calc N/A cm-1
wavenum_min_calc N/A cm-1
----------------------------------------
(0.0, 1.0)
from radis import MergeSlabs, calc_spectrum
spectra = []
for (wmin, wmax) in [(50, 3000), (3000, 7000), (7000, 10000)]:
spectra.append(
calc_spectrum(
wmin,
wmax,
Tgas=1000,
pressure=10, # bar
molecule="OH",
path_length=1,
mole_fraction=0.1,
wstep=0.1,
databank="hitemp",
verbose=False,
)
)
s = MergeSlabs(*spectra, resample="full", out="transparent")
print(s)
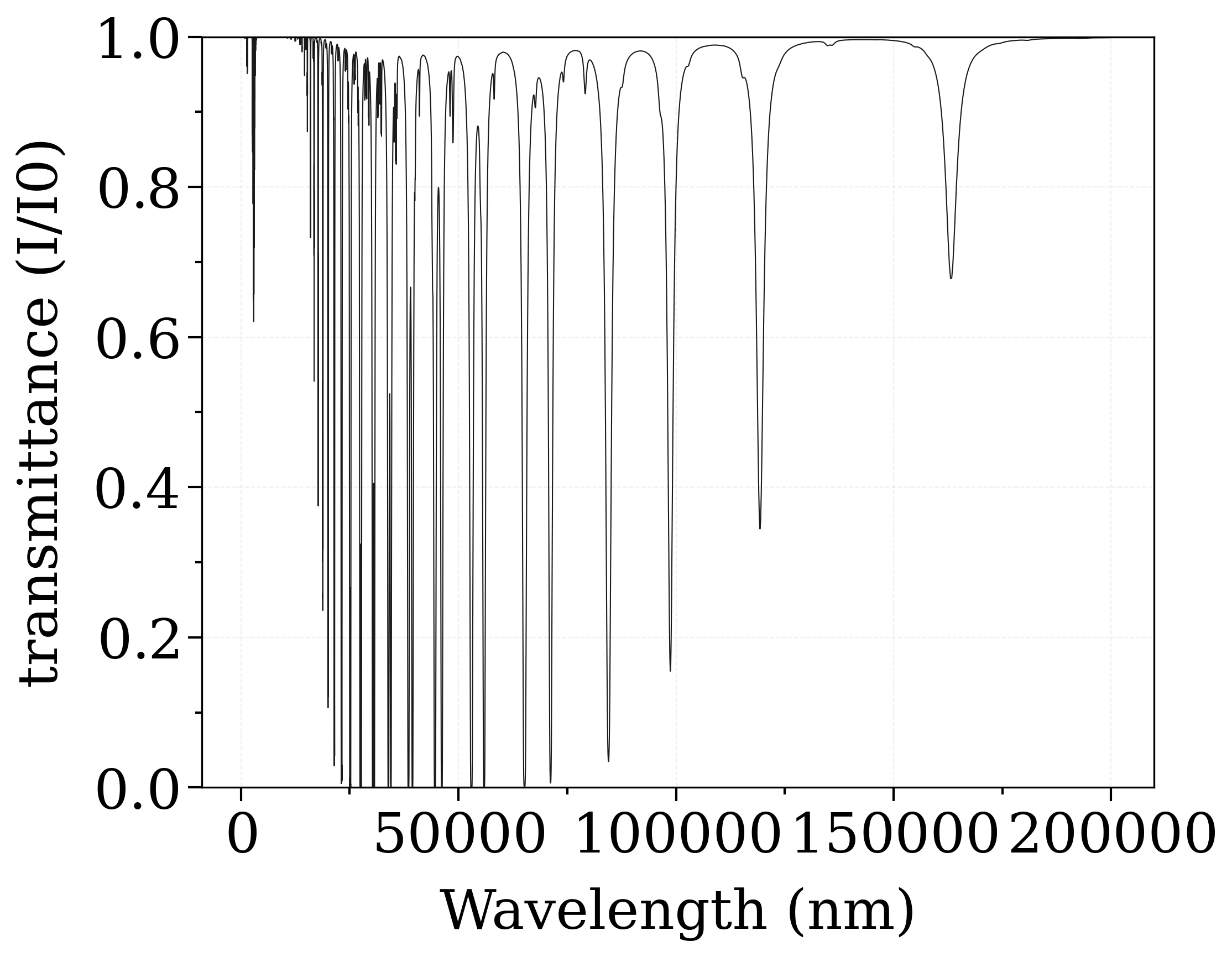
s.plot("transmittance_noslit", wunit="nm")
import matplotlib.pyplot as plt
plt.ylim(0, 1)
Total running time of the script: ( 0 minutes 1.325 seconds)



