Use Custom Abundances¶
Custom isotopologue abundances can be defined, to model non-terrestrial atmospheres.
Abundances are read and set directly in the SpectrumFactory
using the get_abundance()
and set_abundance() methods.
Below, we compute a CO2 spectrum with custom abundances for the two main terrestrial isotopes (12C-16O2 ; 13C-16O2)
See Also¶
from radis.test.utils import setup_test_line_databases
setup_test_line_databases() # creates "HITEMP-CO2-TEST" for this example
from radis import SpectrumFactory
sf = SpectrumFactory(
2284.2,
2284.6,
wstep=0.001, # cm-1
pressure=20 * 1e-3, # bar
mole_fraction=400e-6,
molecule="CO2",
isotope="1,2",
verbose=False,
)
sf.load_databank("HITEMP-CO2-TEST")
Added HITRAN-CO2-TEST database in /home/docs/radis.json
Added HITRAN-CO-TEST database in /home/docs/radis.json
Added HITEMP-CO2-TEST database in /home/docs/radis.json
Added HITEMP-CO2-HAMIL-TEST database in /home/docs/radis.json
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning: The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
df = pd.read_csv(file, comment="#", delim_whitespace=True)
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: MissingReferenceWarning: Missing doi for CDSD-HITEMP. Use HITEMP-2010?
warnings.warn(WarningType(message))
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/tools.py:280: FutureWarning: Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
new_col = df["branch"].replace(mapping)
To explicitly identify the isotopes we can use the molparam attribute the Factory
print(sf.molparam.get("CO2", 1, "isotope_name")) # >> (12C)(16O)2
print(sf.molparam.get("CO2", 2, "isotope_name")) # >> (13C)(16O)2
(12C)(16O)2
(13C)(16O)2
Print the default abundance of the CO2 isotopes, compute a spectrum
print("Abundance of CO2[1,2]", sf.get_abundance("CO2", [1, 2]))
sf.eq_spectrum(2000).plot("abscoeff")
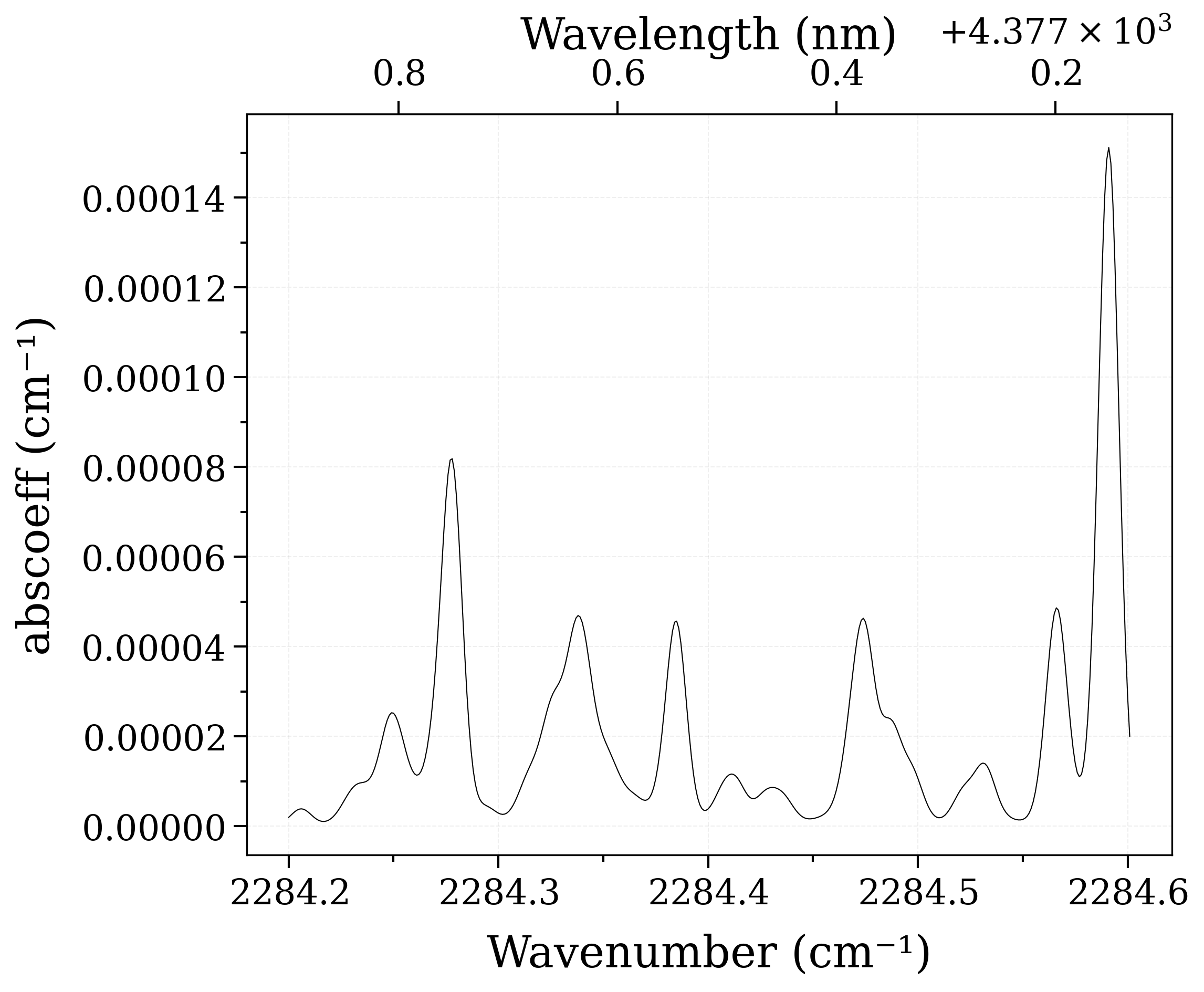
Abundance of CO2[1,2] [[0.984204 ]
[0.0110574]]
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: PerformanceWarning: 'object' type column found in database, calculations and memory usage would be faster with a numeric type. Possible solution is to not use 'save_memory' and convert the columns to dtype.
warnings.warn(WarningType(message))
<matplotlib.lines.Line2D object at 0x7fbdfe9e2830>
Set the abundance of CO2(626) to 0.8; and the abundance of CO2(636) to 0.2 (arbitrary):
sf.set_abundance("CO2", [1, 2], [0.8, 0.2])
print("New abundance of CO2[1,2]", sf.get_abundance("CO2", [1, 2]))
sf.eq_spectrum(2000).plot("abscoeff", nfig="same")
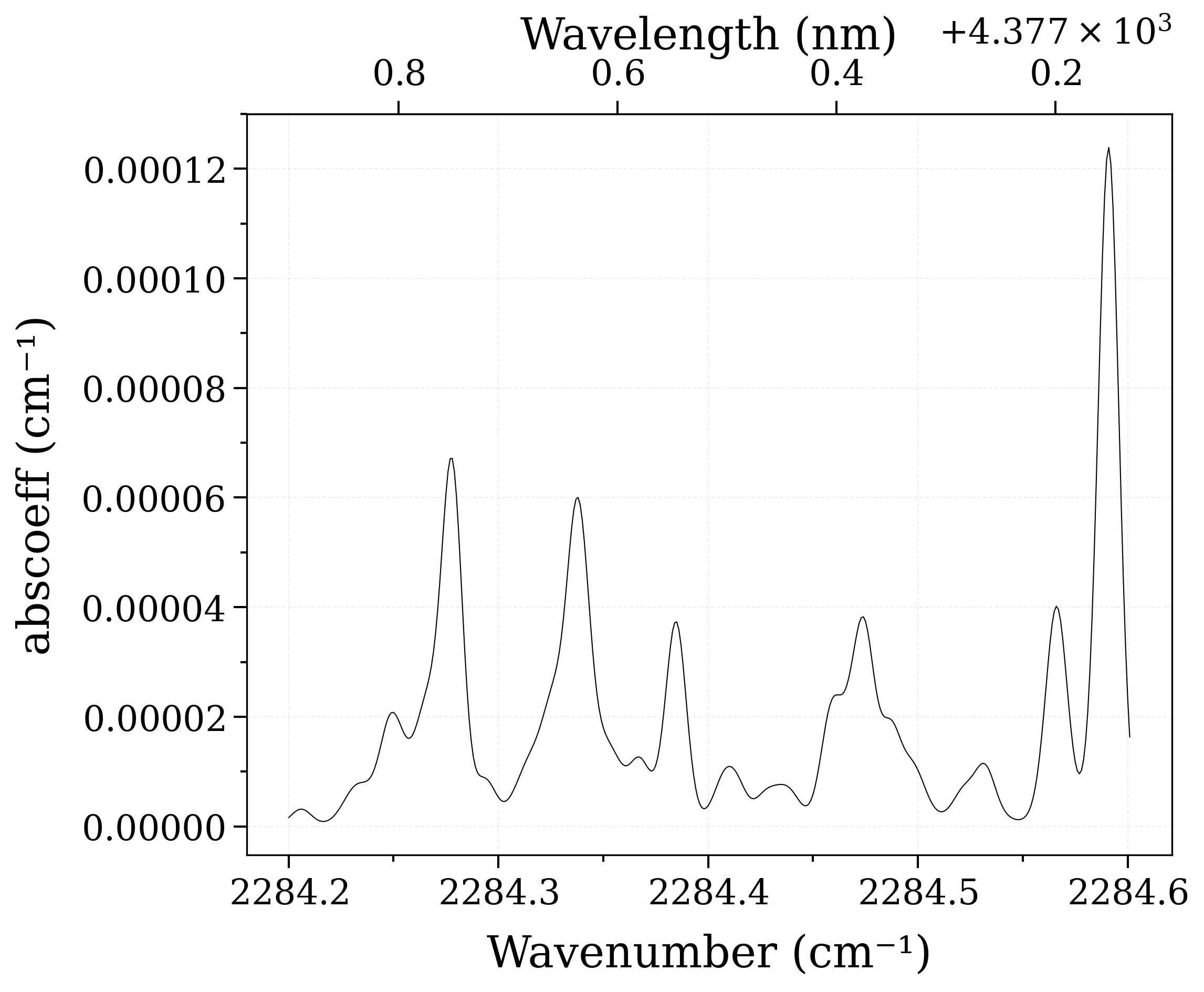
New abundance of CO2[1,2] [[0.8]
[0.2]]
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: PerformanceWarning: 'object' type column found in database, calculations and memory usage would be faster with a numeric type. Possible solution is to not use 'save_memory' and convert the columns to dtype.
warnings.warn(WarningType(message))
findfont: Font family ['sans-serif'] not found. Falling back to DejaVu Sans.
findfont: Generic family 'sans-serif' not found because none of the following families were found: Helvetica, Bitstream Vera Sans, Lucida Grande, Verdana, Geneva, Lucid, Arial, Avant Garde, sans-serif
<matplotlib.lines.Line2D object at 0x7fbdfe79e7d0>
Total running time of the script: (0 minutes 2.189 seconds)



