radis.io.exomol module¶
Created on Sun May 22 18:11:23 2022
@author: erwan
- fetch_exomol(molecule, database=None, local_databases=None, databank_name='EXOMOL-{molecule}', isotope='1', load_wavenum_min=None, load_wavenum_max=None, columns=None, cache=True, verbose=True, clean_cache_files=True, return_local_path=False, return_partition_function=False, engine='default', output='pandas', skip_optional_data=True)[source]¶
Stream ExoMol file from EXOMOL website. Unzip and build a HDF5 file directly.
Returns a Pandas DataFrame containing all lines.
- Parameters:
molecule (
str) – ExoMol moleculedatabase (
str) – database name. Ex:POKAZATELorBT2forH2O. SeeKNOWN_EXOMOL_DATABASE_NAMES. IfNoneand there is only one database available, use it.local_databases (
str) – where to create the RADIS HDF5 files. Default"~/.radisdb/exomol". Can be changed inradis.config["DEFAULT_DOWNLOAD_PATH"]or in ~/radis.json config filedatabank_name (
str) – name of the databank in RADIS Configuration file Default"EXOMOL-{molecule}"isotope (
strorint) – load only certain isotopes, sorted by terrestrial abundances :'1','2', etc. Default1.Note
In RADIS, isotope abundance is included in the line intensity calculation. However, the terrestrial abundances used may not be relevant to non-terrestrial applications. By default, the abundance is given reading HITRAN data. If the molecule does not exist in the HITRAN database, the abundance is read from the
radis/radis_default.jsonconfiguration file, which can be modified by editingradis.configafter import or directly by editing the user~/radis.jsonuser configuration file (overwritesradis_default.json). In theradis/radis_default.jsonfile, values were calculated with a simple model based on the terrestrial isotopic abundance of each element.load_wavenum_min, load_wavenum_max (float (cm-1)) – load only specific wavenumbers.
columns (list of str) – list of columns to load. If
None, returns all columns in the file.
- Other Parameters:
cache (bool, or
'regen'or'force') – ifTrue, use existing HDF5 file. IfFalseor'regen', rebuild it. If'force', crash if not cache file found. DefaultTrue.verbose (bool)
clean_cache_files (bool) – if
Trueclean downloaded cache files after HDF5 are created.return_local_path (bool) – if
True, also returns the path of the local database file.return_partition_function (bool) – if
True, also returns aPartFuncExoMolobject.engine (‘vaex’, ‘feather’) – which memory-mapping library to use. If ‘default’ use the value from ~/radis.json
output (‘pandas’, ‘vaex’, ‘jax’) – format of the output DataFrame. If
'jax', returns a dictionary of jax arrays. If'vaex', output is avaex.dataframe.DataFrameLocalNote
Vaex DataFrames are memory-mapped. They do not take any space in RAM and are extremely useful to deal with the largest databases.
skip_optional_data (bool) – If False, fetch all fields which are marked as available in the ExoMol definition file. If True, load only the first 4 columns of the states file (“i”, “E”, “g”, “J”). The structure of the columns above 5 depend on the the definitions file (*.def) and the Exomol version. If
skip_optional_data=False, two errors may occur:a field is marked as present/absent in the *.def field but is absent/present in the *.states file (ie both files are inconsistent).
in the updated version of Exomol, new fields have been added in the states file of some species. But it has not been done for all species, so both structures exist. For instance, the states file of https://exomol.com/data/molecules/HCl/1H-35Cl/HITRAN-HCl/ follows the structure described in [1], unlike the states file of https://exomol.com/data/molecules/NO/14N-16O/XABC/ which follows the structure described in [2].
- Returns:
df (pd.DataFrame or vaex.dataframe.DataFrameLocal) – Line list A HDF5 file is also created in
local_databasesand referenced in the RADIS config file with namedatabank_namelocal_path (str) – path of local database file if
return_local_path
Examples
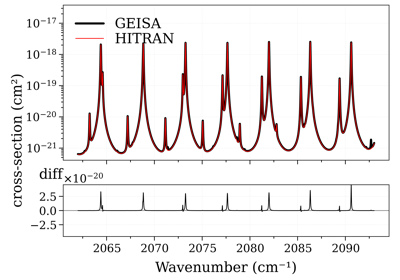
Compare CO cross-sections from HITRAN, HITEMP, GEISA, and ExoMol
Compare CO cross-sections from HITRAN, HITEMP, GEISA, and ExoMolNotes
if using
load_only_wavenum_above/beloworisotope, the whole database is anyway downloaded and uncompressed tolocal_databasesfast access .HDF5 files (which will take a long time on first call). Only the expected wavenumber range & isotopes are returned. The .HFD5 parsing useshdf2df()References
See also