Compare CO cross-sections from HITRAN, HITEMP, GEISA, and ExoMol¶
Auto-download and calculate CO spectrum from the HITRAN, HITEMP, GEISA, and ExoMol databases.
ExoMol references multiple databases for CO. Here we do not
use the ExoMol recommended database (see get_exomol_database_list())
but we use the HITEMP database hosted on ExoMol.
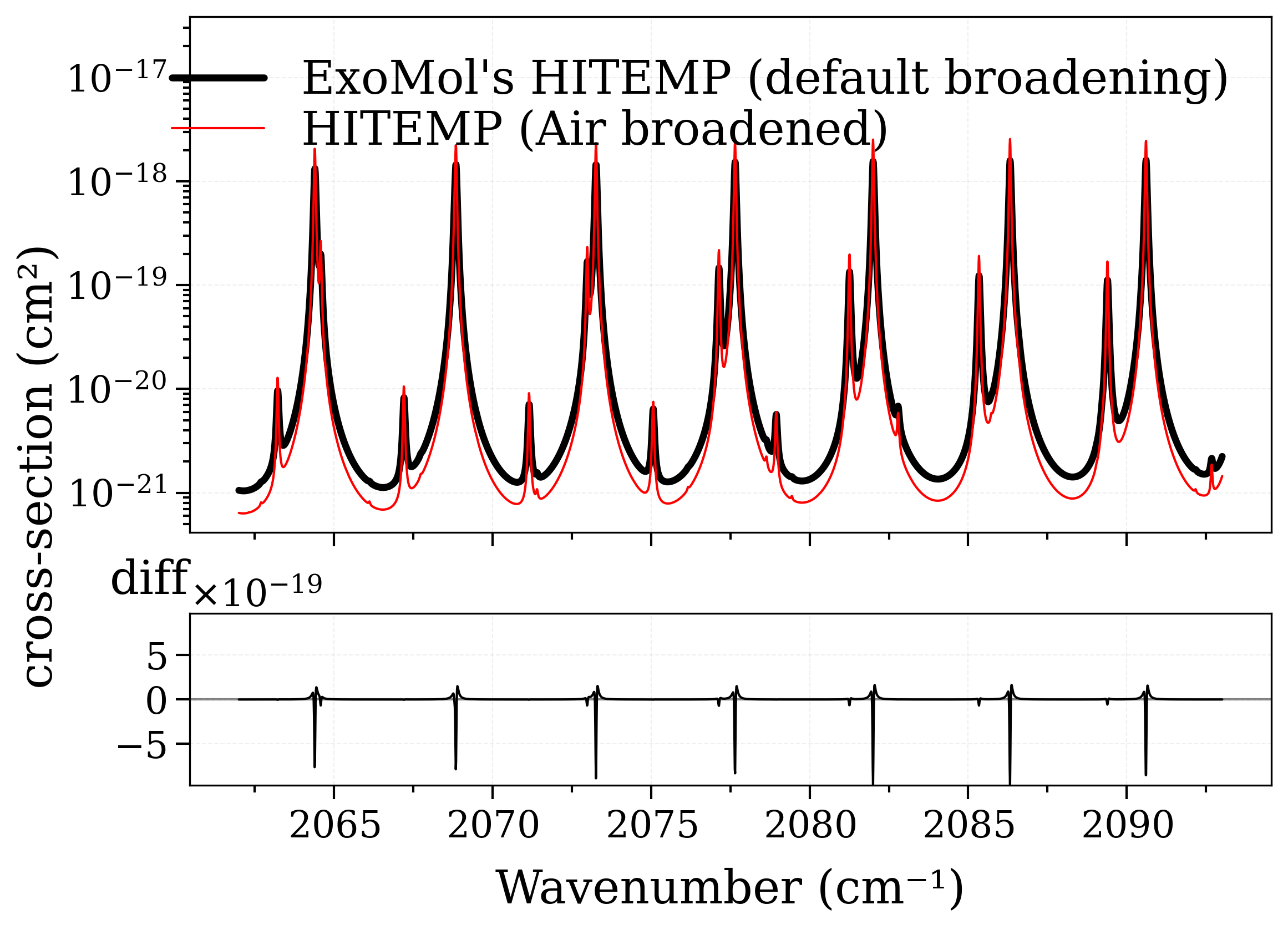
Output should be similar, but no! By default these ExoMol does not provide broadening coefficients for air. However, the Einstein coefficients & linestrengths should be the same, therefore the integrals under the lines should be similar.
We verify this below :
For further exploration, ExoMol and HITEMP lines can be downloaded and accessed separately using
fetch_exomol() and fetch_hitemp()
import astropy.units as u
from radis import calc_spectrum, plot_diff
conditions = {
"wmin": 2062 / u.cm,
"wmax": 2093 / u.cm,
"molecule": "CO",
"isotope": "1",
"pressure": 1.01325, # bar
"Tgas": 1000, # K
"mole_fraction": 0.1,
"path_length": 1, # cm
"verbose": True,
"neighbour_lines": 20, # we account for the effect on neighbour_lines by computing ``20cm-1``
}
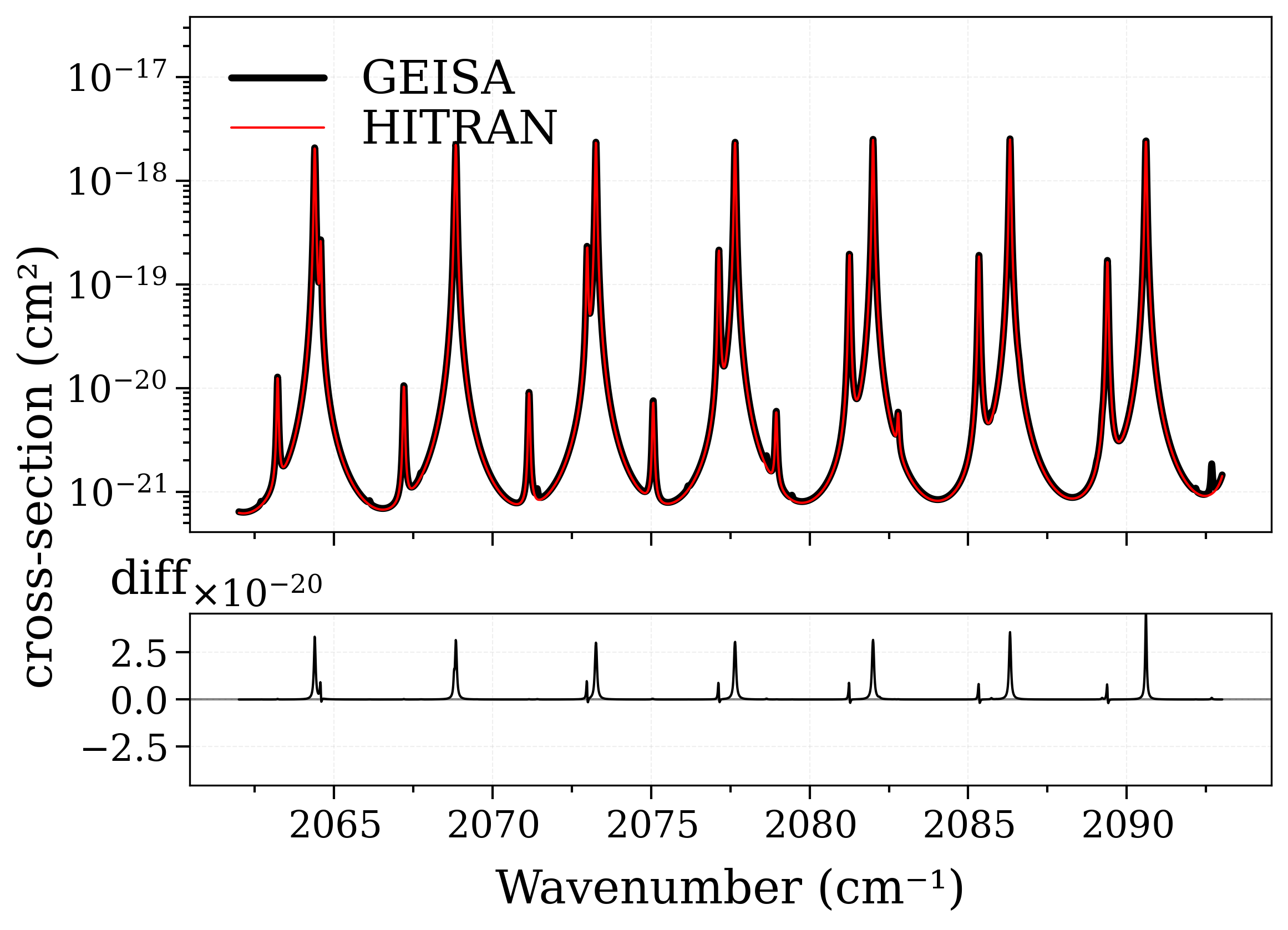
s_geisa = calc_spectrum(**conditions, databank="geisa", name="GEISA")
s_hitran = calc_spectrum(**conditions, databank="hitran", name="HITRAN")
fig, [ax0, ax1] = plot_diff(s_geisa, s_hitran, "xsection", yscale="log")
# Adjust diff plot to be in linear scale
ax1.set_yscale("linear")
ax0.set_ylim(ymax=ax0.get_ylim()[1] * 10) # more space for legend
# Note: these two spectra are alike.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
Molecule: CO
Downloading line_GEISA2020_asc_gs08_v1.0_co for CO (1/1).
Added GEISA-CO database in /home/docs/radis.json
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2093.0000 cm-1
wavenum_min 2062.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat geisa
dbpath /home/docs/.radisdb/geisa/CO-line_GEISA2020_asc_gs08_v1.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 20 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 7102
----------------------------------------
0.03s - Spectrum calculated
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
Using /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO
Data is fetched from http://hitran.org
BEGIN DOWNLOAD: CO_1
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_1.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_1.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_1.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_1.data
Header written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_1.header
END DOWNLOAD
Lines parsed: 1344
PROCESSED
Data is fetched from http://hitran.org
BEGIN DOWNLOAD: CO_2
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_2.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_2.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_2.data
Header written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_2.header
END DOWNLOAD
Lines parsed: 1042
PROCESSED
Data is fetched from http://hitran.org
BEGIN DOWNLOAD: CO_3
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_3.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_3.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_3.data
Header written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_3.header
END DOWNLOAD
Lines parsed: 920
PROCESSED
Data is fetched from http://hitran.org
BEGIN DOWNLOAD: CO_4
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_4.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_4.data
Header written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_4.header
END DOWNLOAD
Lines parsed: 800
PROCESSED
Data is fetched from http://hitran.org
BEGIN DOWNLOAD: CO_5
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_5.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_5.data
Header written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_5.header
END DOWNLOAD
Lines parsed: 674
PROCESSED
Data is fetched from http://hitran.org
BEGIN DOWNLOAD: CO_6
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_6.data
65536 bytes written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_6.data
Header written to /home/docs/.radisdb/hitran/downloads__can_be_deleted/CO/CO_6.header
END DOWNLOAD
Lines parsed: 601
PROCESSED
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/tools.py:280: FutureWarning:
Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/tools.py:280: FutureWarning:
Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/tools.py:280: FutureWarning:
Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/tools.py:280: FutureWarning:
Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/tools.py:280: FutureWarning:
Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/hitranapi.py:128: FutureWarning:
A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/tools.py:280: FutureWarning:
Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
Added HITRAN-CO database in /home/docs/radis.json
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: HighTemperatureWarning:
HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2093.0000 cm-1
wavenum_min 2062.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/.radisdb/hitran/CO.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 20 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 7102
----------------------------------------
0.03s - Spectrum calculated
(4.083380058233573e-22, 3.843485051808716e-17)
s_exomol = calc_spectrum(
**conditions, databank="exomol", name="ExoMol's HITEMP (default broadening)"
)
s_hitemp = calc_spectrum(
**conditions,
databank="hitemp",
name="HITEMP (Air broadened)",
)
fig, [ax0, ax1] = plot_diff(s_exomol, s_hitemp, "xsection", yscale="log")
# Adjust diff plot to be in linear scale
ax1.set_yscale("linear")
ax0.set_ylim(ymax=ax0.get_ylim()[1] * 10) # more space for legend
# Broadening coefficients are different in these databases, so lineshapes
# end up being very different; however the areas under the lines should be the same.
# We verify this :
print(
f"Ratio of integrated area = {s_exomol.get_integral('xsection')/s_hitemp.get_integral('xsection'):.2f}"
)
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
Using the recommended ExoMol databases for CO
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
========== Loading Exomol database [start] ==========
=> Downloading from http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.def
=> Downloading from http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.pf
=> Downloading from http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.states.bz2
=> Downloading from http://www.exomol.com/db/CO/12C-16O/12C-16O__H2.broad
=> Downloading from http://www.exomol.com/db/CO/12C-16O/12C-16O__He.broad
=> Downloading from http://www.exomol.com/db/CO/12C-16O/12C-16O__air.broad
Error: Couldn't download .broad file at http://www.exomol.com/db/CO/12C-16O/12C-16O__air.broad and save.
Note: Caching states data to the vaex format. After the second time, it will become much faster.
Molecule: CO
Isotopologue: 12C-16O
Background atmosphere: Air
ExoMol database: Li2015
Local folder: /home/docs/.radisdb/exomol/CO/12C-16O/Li2015
Transition files:
=> File 12C-16O__Li2015.trans
=> Downloading from http://www.exomol.com/db/CO/12C-16O/Li2015/12C-16O__Li2015.trans.bz2
=> Caching the *.trans.bz2 file to the vaex (*.h5) format. After the second time, it will become much faster.
=> You can deleted the 'trans.bz2' file by hand.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/api/exomolapi.py:1258: UserWarning:
Could not load `12C-16O__Air.broad`. The default broadening parameters are used.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
========== Loading Exomol database [end] ==========
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/misc/warning.py:427: MissingPressureShiftWarning:
Pressure-shift coefficient not given in database: assumed 0 pressure shift
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2093.0000 cm-1
wavenum_min 2062.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat exomol-radisdb
dbpath /home/docs/.radisdb/exomol/CO/12C-16O/Li2015
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 20 cm-1
optimization simple
parfuncfmt exomol
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 7102
----------------------------------------
0.02s - Spectrum calculated
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/latest/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2093.0000 cm-1
wavenum_min 2062.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitemp-radisdb
dbpath /home/docs/.radisdb/hitemp/CO-05_HITEMP2019.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 20 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 7102
----------------------------------------
0.03s - Spectrum calculated
Ratio of integrated area = 1.00
Total running time of the script: (0 minutes 13.325 seconds)



