radis.io.hitran module¶
Summary¶
HITRAN database parser
Routine Listing¶
- class HITRANDatabaseManager(name, molecule, local_databases, engine='default', extra_params=None, verbose=True, parallel=True)[source]¶
Bases:
DatabaseManager- download_and_parse(local_file, cache=True, parse_quanta=True)[source]¶
Download from HITRAN and parse into
local_file. Also add metadataOverwrites
radis.io.dbmanager.DatabaseManager.download_and_parse()which downloads from a list of URL, because here we use [HAPI] to download the files.- Parameters
opener (an opener with an .open() command)
gfile (file handler. Filename: for info)
- cast_to_int64_with_missing_values(dg, keys)[source]¶
replace missing values of int64 columns with -1
- columns_2004 = {'A': ('a10', <class 'float'>, 'Einstein A coefficient', 's-1'), 'El': ('a10', <class 'float'>, 'lower-state energy', 'cm-1'), 'Pshft': ('a8', <class 'float'>, 'air pressure-induced line shift at 296K', 'cm-1.atm-1'), 'Tdpair': ('a4', <class 'float'>, 'temperature-dependance exponent for Gamma air', ''), 'airbrd': ('a5', <class 'float'>, 'air-broadened half-width at 296K', 'cm-1.atm-1'), 'globl': ('a15', <class 'str'>, 'electronic and vibrational global lower quanta', ''), 'globu': ('a15', <class 'str'>, 'electronic and vibrational global upper quanta', ''), 'gp': ('a7', <class 'float'>, 'upper state degeneracy', ''), 'gpp': ('a7', <class 'float'>, 'lower state degeneracy', ''), 'id': ('a2', <class 'int'>, 'Molecular number', ''), 'ierr': ('a6', <class 'str'>, 'ordered list of indices corresponding to uncertainty estimates of transition parameters', ''), 'int': ('a10', <class 'float'>, 'intensity at 296K', 'cm-1/(molecule/cm-2)'), 'iref': ('a12', <class 'str'>, 'ordered list of reference identifiers for transition parameters', ''), 'iso': ('a1', <class 'int'>, 'isotope number', ''), 'lmix': ('a1', <class 'str'>, 'flag indicating the presence of additional data and code relating to line-mixing', ''), 'locl': ('a15', <class 'str'>, 'electronic and vibrational local lower quanta', ''), 'locu': ('a15', <class 'str'>, 'electronic and vibrational local upper quanta', ''), 'selbrd': ('a5', <class 'float'>, 'self-broadened half-width at 296K', 'cm-1.atm-1'), 'wav': ('a12', <class 'float'>, 'vacuum wavenumber', 'cm-1')}[source]¶
parsing order of HITRAN 2004 format
- Type
OrderedDict
- fetch_hitran(molecule, extra_params=None, local_databases=None, databank_name='HITRAN-{molecule}', isotope=None, load_wavenum_min=None, load_wavenum_max=None, columns=None, cache=True, verbose=True, clean_cache_files=True, return_local_path=False, engine='default', output='pandas', parallel=True, parse_quanta=True)[source]¶
Download all HITRAN lines from HITRAN website. Unzip and build a HDF5 file directly.
Returns a Pandas DataFrame containing all lines.
- Parameters
molecule (str) – one specific molecule name, listed in HITRAN molecule metadata. See https://hitran.org/docs/molec-meta/ Example: “H2O”, “CO2”, etc.
local_databases (str) – where to create the RADIS HDF5 files. Default
"~/.radisdb/hitran". Can be changed inradis.config["DEFAULT_DOWNLOAD_PATH"]or in ~/radis.json config filedatabank_name (str) – name of the databank in RADIS Configuration file Default
"HITRAN-{molecule}"isotope (str) – load only certain isotopes :
'2','1,2', etc. IfNone, loads everything. DefaultNone.load_wavenum_min, load_wavenum_max (float (cm-1)) – load only specific wavenumbers.
columns (list of str) – list of columns to load. If
None, returns all columns in the file.extra_params (‘all’ or None) – Downloads all additional columns available in the HAPI database for the molecule including parameters like
gamma_co2,n_co2that are required to calculate spectrum in co2 diluent. For eg:from radis.io.hitran import fetch_hitran df = fetch_hitran('CO', extra_params='all', cache='regen') # cache='regen' to regenerate new database with additional columns
- Other Parameters
cache (
True,False,'regen'or'force') – ifTrue, use existing HDF5 file. IfFalseor'regen', rebuild it. If'force', raise an error if cache file cannot be used (useful for debugging). DefaultTrue.verbose (bool)
clean_cache_files (bool) – if
Trueclean downloaded cache files after HDF5 are created.return_local_path (bool) – if
True, also returns the path of the local database file.engine (‘pytables’, ‘vaex’, ‘default’) – which HDF5 library to use. If ‘default’ use the value from ~/radis.json
output (‘pandas’, ‘vaex’, ‘jax’) – format of the output DataFrame. If
'jax', returns a dictionary of jax arrays. If'vaex', output is avaex.dataframe.DataFrameLocalNote
Vaex DataFrames are memory-mapped. They do not take any space in RAM and are extremelly useful to deal with the largest databases.
parallel (bool) – if
True, uses joblib.parallel to load database with multiple processesparse_quanta (bool) – if
True, parse local & global quanta (required to identify lines for non-LTE calculations ; but sometimes lines are not labelled.)
- Returns
df (pd.DataFrame) – Line list A HDF5 file is also created in
local_databasesand referenced in the RADIS config file with namedatabank_namelocal_path (str) – path of local database file if
return_local_path
Examples
from radis.io.hitran import fetch_hitran df = fetch_hitran("CO") print(df.columns) >>> Index(['id', 'iso', 'wav', 'int', 'A', 'airbrd', 'selbrd', 'El', 'Tdpair', 'Pshft', 'gp', 'gpp', 'branch', 'jl', 'vu', 'vl'], dtype='object')
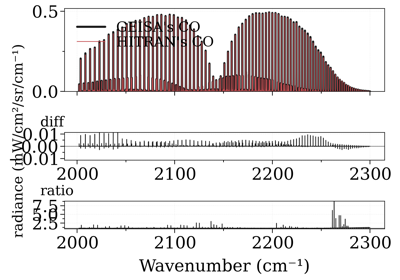
Compare CO spectrum from the GEISA and HITRAN database
Compare CO spectrum from the GEISA and HITRAN databaseNotes
if using
load_only_wavenum_above/beloworisotope, the whole database is anyway downloaded and uncompressed tolocal_databasesfast access .HDF5 files (which will take a long time on first call). Only the expected wavenumber range & isotopes are returned. The .HFD5 parsing useshdf2df()See also
- hit2df(fname, cache=True, verbose=True, drop_non_numeric=True, load_wavenum_min=None, load_wavenum_max=None, engine='pytables', parse_quanta=True)[source]¶
Convert a HITRAN/HITEMP [1]_ file to a Pandas dataframe
- Parameters
fname (str) – HITRAN-HITEMP file name
cache (boolean, or
'regen'or'force') – ifTrue, a pandas-readable HDF5 file is generated on first access, and later used. This saves on the datatype cast and conversion and improves performances a lot (but changes in the database are not taken into account). If False, no database is used. If'regen', temp file are reconstructed. DefaultTrue.
- Other Parameters
drop_non_numeric (boolean) – if
True, non numeric columns are dropped. This improves performances, but make sure all the columns you need are converted to numeric formats before hand. DefaultTrue. Note that if a cache file is loaded it will be left untouched.load_wavenum_min, load_wavenum_max (float) – if not
'None', only load the cached file if it contains data for wavenumbers above/below the specified value. See :py:func`~radis.io.cache_files.load_h5_cache_file`. Default'None'.engine (‘pytables’, ‘vaex’) – format for Hdf5 cache file. Default
pytablesparse_quanta (bool) – if
True, parse local & global quanta (required to identify lines for non-LTE calculations ; but sometimes lines are not labelled.)
- Returns
df – dataframe containing all lines and parameters
- Return type
pandas Dataframe
References
Notes
Performances: see CDSD-HITEMP parser
See also
- parse_global_quanta(df, mol, verbose=True)[source]¶
- Parameters
df (pandas Dataframe)
mol (str) – molecule name
- parse_local_quanta(df, mol, verbose=True)[source]¶
- Parameters
df (pandas Dataframe)
mol (str) – molecule name
- post_process_hitran_data(df, molecule, verbose=True, drop_non_numeric=True, parse_quanta=True)[source]¶
Parsing non-equilibrum parameters in HITRAN/HITEMP [1]_ file to and return final Pandas Dataframe
- Parameters
df (pandas Dataframe) – dataframe containing generic parameters
molecule (str) – molecule name
- Other Parameters
drop_non_numeric (boolean) – if
True, non numeric columns are dropped. This improves performances, but make sure all the columns you need are converted to numeric formats before hand. DefaultTrue. Note that if a cache file is loaded it will be left untouched.parse_quanta (bool) – if
True, parse local & global quanta (required to identify lines for non-LTE calculations ; but sometimes lines are not labelled.)
- Returns
df – dataframe containing all lines and parameters
- Return type
pandas Dataframe
References
Notes
Performances: see CDSD-HITEMP parser
See also



