Post-process using Specutils¶
Find peaks or uncertainties using the specutils library. A Radis Spectrum
object can easily be converted to a specutils specutils.spectra.spectrum1d.Spectrum1D
using to_specutils().
Below, we create a noisy spectrum based on a synthetic CO spectrum,
we convert it to specutils, add uncertainties by targeting a
noisy region, then determine the lines using find_lines_threshold() :
import astropy.units as u
import numpy as np
from radis import test_spectrum
""" We create a synthetic CO spectrum"""
s = (
test_spectrum(molecule="CO", wavenum_min=2000, wavenum_max=2030)
.apply_slit(1.5, "nm")
.take("radiance")
)
s.trim() # removes nans created by the slit convolution boundary effects
noise = np.random.normal(0.0, s.max().value * 0.03, len(s))
s_exp = s + noise
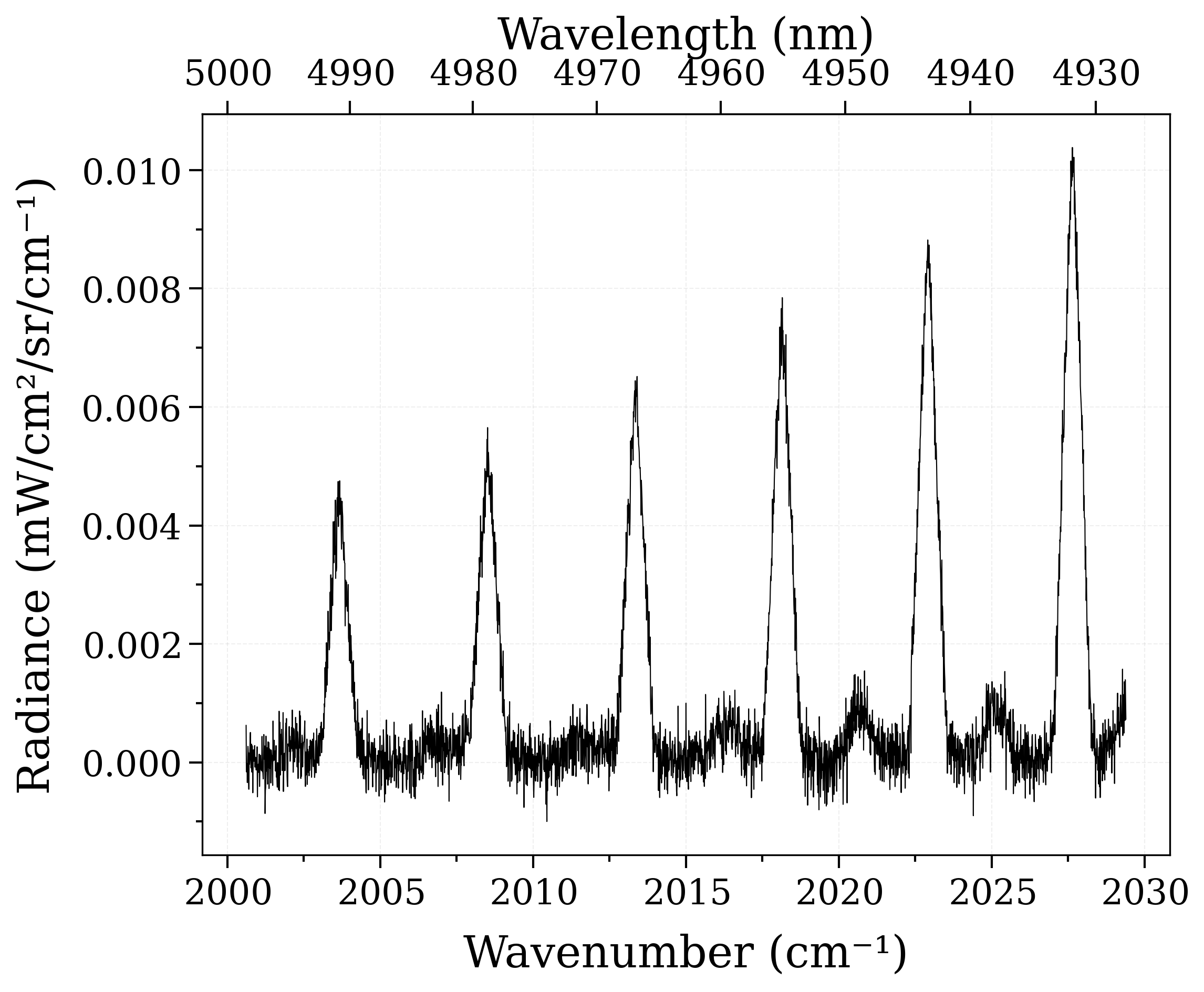
s_exp.plot()
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 700 K
Trot 700 K
Tvib 700 K
isotope 1,2,3
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2030.0000 cm-1
wavenum_min 2000.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/.radisdb/hitran/CO.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 3001
----------------------------------------
0.03s - Spectrum calculated
<matplotlib.lines.Line2D object at 0x7f30cd9dfdf0>
Determine the noise level by selecting a noisy region from the graph above :
spectrum = s_exp.to_specutils()
from specutils import SpectralRegion
from specutils.manipulation import noise_region_uncertainty
noise_region = SpectralRegion(2010.5 / u.cm, 2009.5 / u.cm)
spectrum = noise_region_uncertainty(spectrum, noise_region)
Find lines :
from specutils.fitting import find_lines_threshold
lines = find_lines_threshold(spectrum, noise_factor=2)
print(lines)
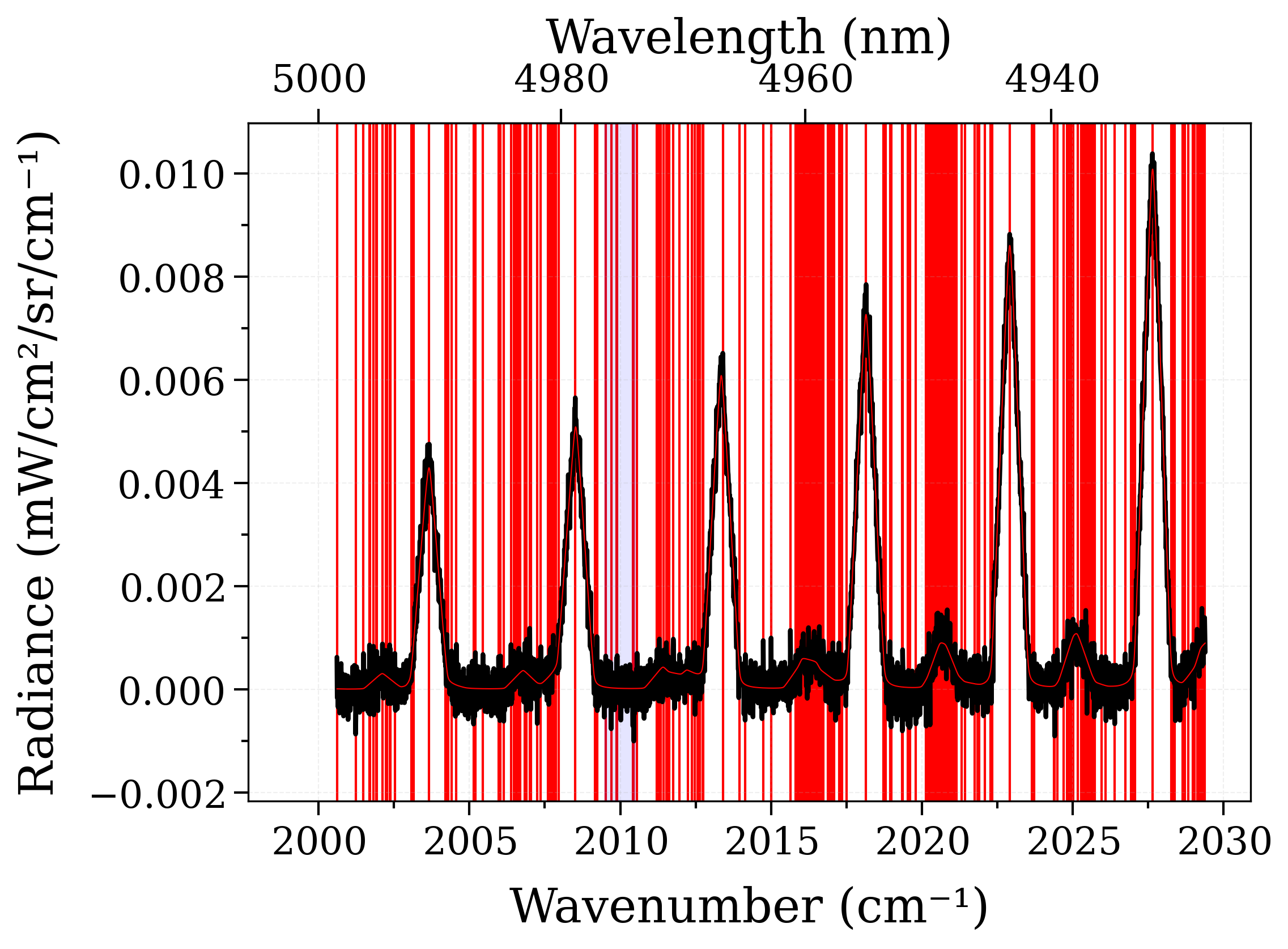
s_exp.plot(lw=2, show_ruler=True)
import matplotlib.pyplot as plt
for line in lines.to_pandas().line_center.values:
plt.axvline(line, color="r", zorder=-1)
s.plot(nfig="same")
plt.axvspan(noise_region.lower.value, noise_region.upper.value, color="b", alpha=0.1)
/home/docs/checkouts/readthedocs.org/user_builds/radis/conda/develop/lib/python3.10/site-packages/specutils/analysis/flux.py:289: AstropyUserWarning:
Spectrum is not below the threshold signal-to-noise 0.01. This may indicate you have not continuum subtracted this spectrum (or that you have but it has high SNR features).
If you want to suppress this warning either type 'specutils.conf.do_continuum_function_check = False' or see http://docs.astropy.org/en/stable/config/#adding-new-configuration-items for other ways to configure the warning.
line_center line_type line_center_index
1 / cm
------------------ ---------- -----------------
2000.6099999999994 emission 0
2001.4899999999986 emission 88
2001.6899999999985 emission 108
2001.7099999999984 emission 110
2001.8199999999983 emission 121
2001.9099999999983 emission 130
2001.9299999999982 emission 132
2002.119999999998 emission 151
2002.229999999998 emission 162
2002.269999999998 emission 166
... ... ...
2018.9799999999827 absorption 1837
2019.3499999999824 absorption 1874
2019.5399999999822 absorption 1893
2019.5899999999822 absorption 1898
2019.6099999999822 absorption 1900
2019.799999999982 absorption 1919
2020.1399999999817 absorption 1953
2020.2699999999816 absorption 1966
2024.3999999999778 absorption 2379
2026.389999999976 absorption 2578
Length = 216 rows
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/tools/plot_tools.py:619: UserWarning:
Couldn't add Ruler tool (still an experimental feature in RADIS : please report the error !)
<matplotlib.patches.Polygon object at 0x7f30c1f35390>
Note: we can also create a RADIS spectrum object from Specutils
specutils.spectra.spectrum1d.Spectrum1D :
from radis import Spectrum
s2 = Spectrum.from_specutils(spectrum)
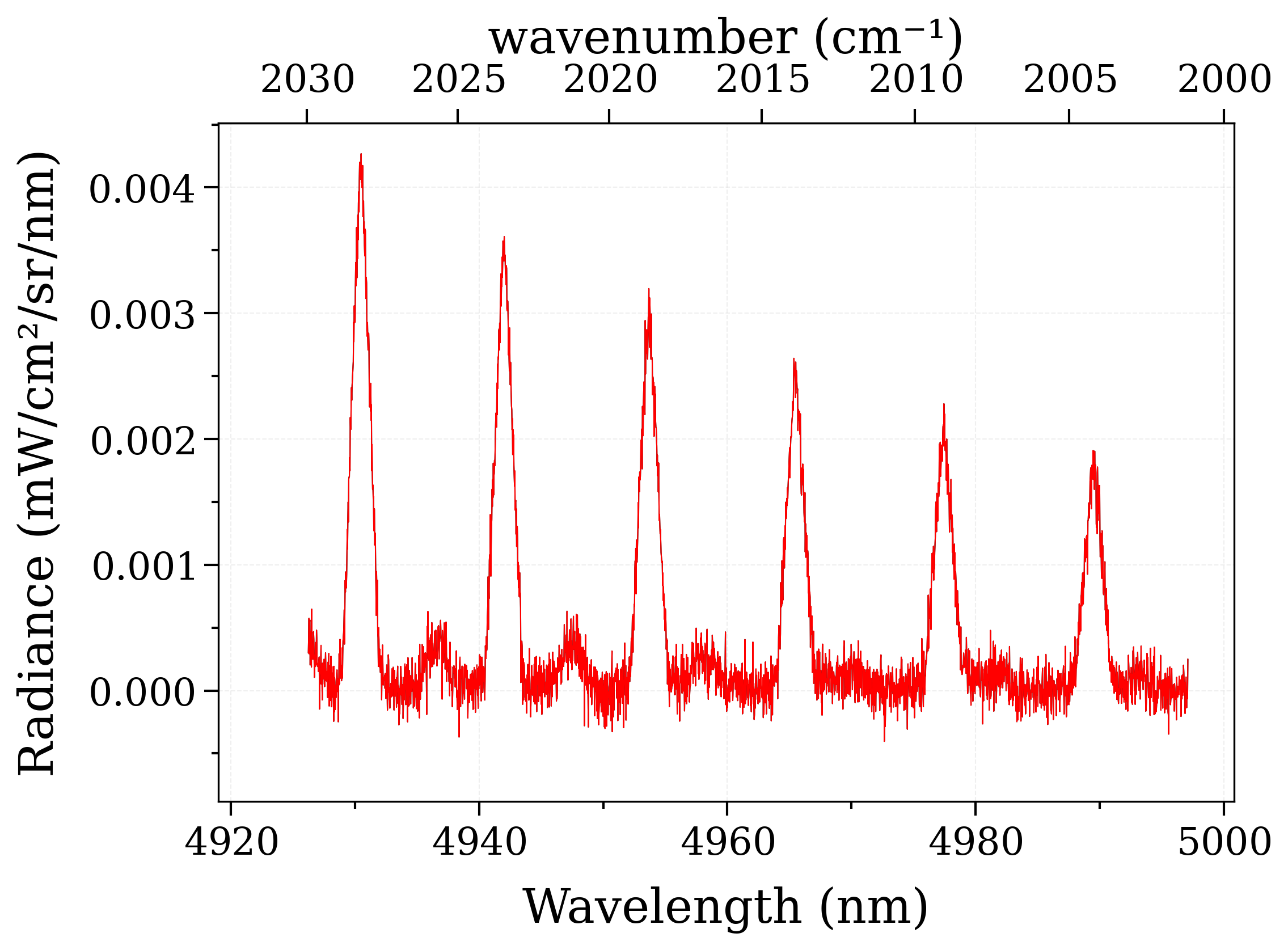
s2.plot(Iunit="mW/cm2/sr/nm", wunit="nm")
s_exp.plot(Iunit="mW/cm2/sr/nm", wunit="nm", nfig="same")
assert s_exp == s2
Total running time of the script: (0 minutes 2.995 seconds)



