Calculate non-LTE spectra of carbon-monoxide¶
Compute a CO spectrum with the temperature of the vibrational mode different from the temperature of the rotational mode.
This example uses the calc_spectrum() function,
the [HITRAN-2016] line database to derive the line positions
and intensities, and the default RADIS spectroscopic
constants to compute nonequilibrium energies and populations,
but it can be extended to other line databases and other sets
of spectroscopic constants.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
Calculating energy levels with Dunham expansion for CO(X1Σ+)(iso1)
Database generated up to v=48, J=238
Calculating energy levels with Dunham expansion for CO(X1Σ+)(iso2)
Database generated up to v=48, J=243
Calculating energy levels with Dunham expansion for CO(X1Σ+)(iso3)
Database generated up to v=48, J=243
Calculating Non-Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 300.0 K
Trot 300.0 K
Tvib 700.0 K
isotope 1,2,3
mole_fraction 0.1
molecule CO
path_length 1.0 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2300.0000 cm-1
wavenum_min 1900.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/.radisdb/hitran/CO.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm auto
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding -1
----------------------------------------
Fetching Evib & Erot.
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/misc/warning.py:427: NegativeEnergiesWarning:
There are negative rotational energies in the database
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/misc/warning.py:427: PerformanceWarning:
'gu' was recomputed although 'gp' already in DataFrame. All values are equal
... sorting lines by vibrational bands
... lines sorted in 0.0s
Index(['Tdpair', 'A', 'jl', 'vl', 'int', 'iso', 'wav', 'airbrd', 'vu',
'selbrd', 'gp', 'branch', 'El', 'Pshft', 'ju', 'Eu', 'Evibl', 'Evibu',
'Erotu', 'Erotl', 'gju', 'gjl', 'grotu', 'grotl', 'gvibu', 'gvibl',
'gu', 'gl', 'Rs2', 'Blu', 'Bul', 'Aul', 'viblvl_l', 'viblvl_u', 'band'],
dtype='object')
Index(['Tdpair', 'A', 'jl', 'vl', 'int', 'iso', 'wav', 'airbrd', 'vu',
'selbrd', 'gp', 'branch', 'El', 'Pshft', 'ju', 'Eu', 'Evibl', 'Evibu',
'Erotu', 'Erotl', 'gju', 'gjl', 'grotu', 'grotl', 'gvibu', 'gvibl',
'gu', 'gl', 'Rs2', 'Blu', 'Bul', 'Aul', 'viblvl_l', 'viblvl_u', 'band',
'Qvib', 'Q', 'Qrotl', 'Qrotu'],
dtype='object')
Index(['Tdpair', 'A', 'jl', 'vl', 'int', 'iso', 'wav', 'airbrd', 'vu',
'selbrd', 'gp', 'branch', 'El', 'Pshft', 'ju', 'Eu', 'Evibl', 'Evibu',
'Erotu', 'Erotl', 'gju', 'gjl', 'grotu', 'grotl', 'gvibu', 'gvibl',
'gu', 'gl', 'Rs2', 'Blu', 'Bul', 'Aul', 'viblvl_l', 'viblvl_u', 'band',
'Qvib', 'Q', 'Qrotl', 'Qrotu'],
dtype='object')
0.20s - Spectrum calculated
<matplotlib.lines.Line2D object at 0x7f30b1f83a30>
from astropy import units as u
from radis import calc_spectrum
s2 = calc_spectrum(
1900 / u.cm,
2300 / u.cm,
molecule="CO",
isotope="1,2,3",
pressure=1.01325 * u.bar,
Tvib=700 * u.K,
Trot=300 * u.K,
mole_fraction=0.1,
path_length=1 * u.cm,
databank="hitran", # or use 'hitemp'
)
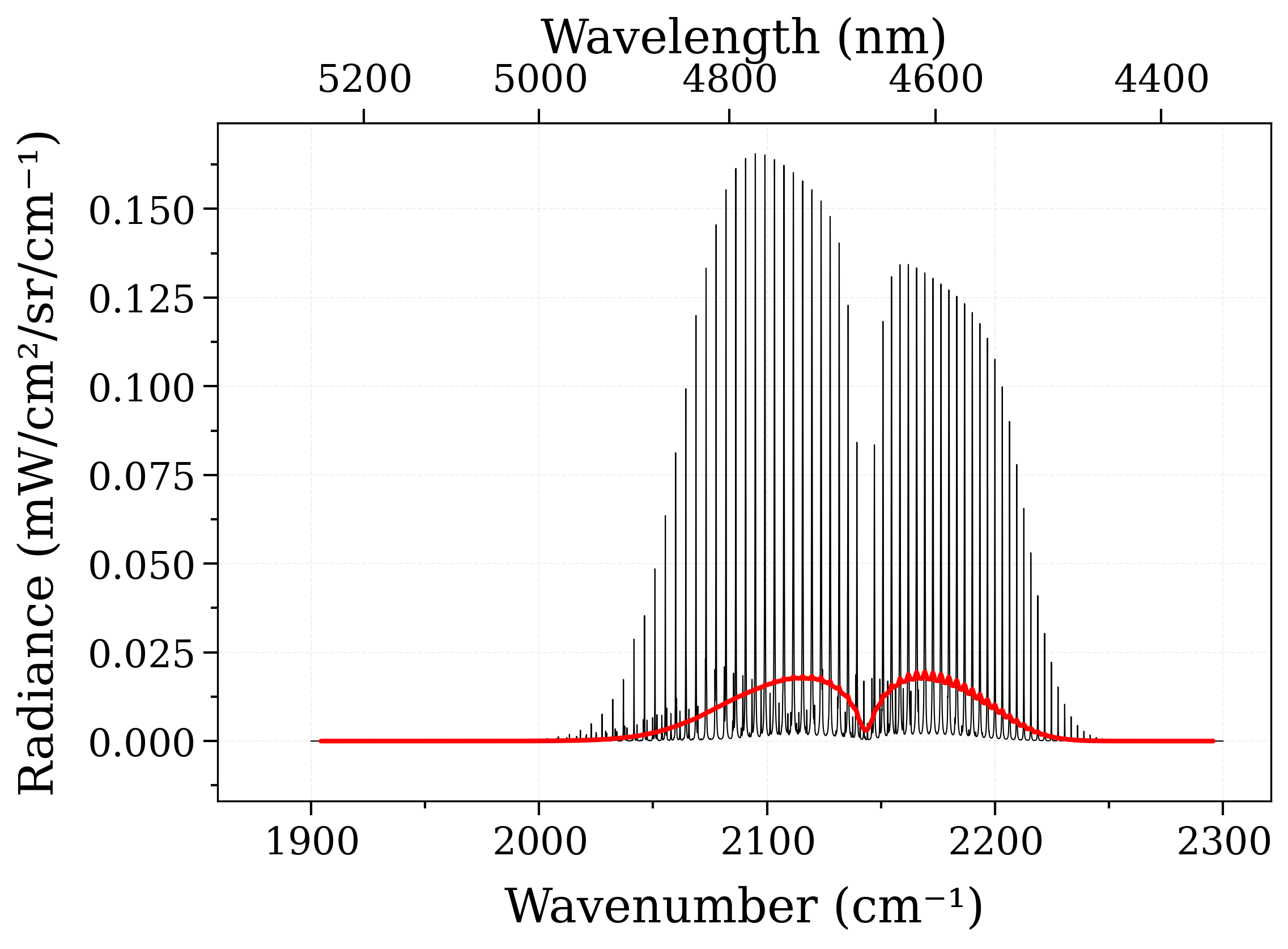
s2.plot("radiance_noslit")
# Apply a (large) instrumental slit function :
s2.apply_slit(10, "nm")
s2.plot("radiance", nfig="same", lw=2) # compare with previous
Total running time of the script: (0 minutes 2.163 seconds)



