Compare CO spectrum from the GEISA and HITRAN database¶
GEISA Database has been newly implemented in RADIS 0.13 release on May 15, 2022. This is among the very first attempts to compare the spectra generated from the two databases.
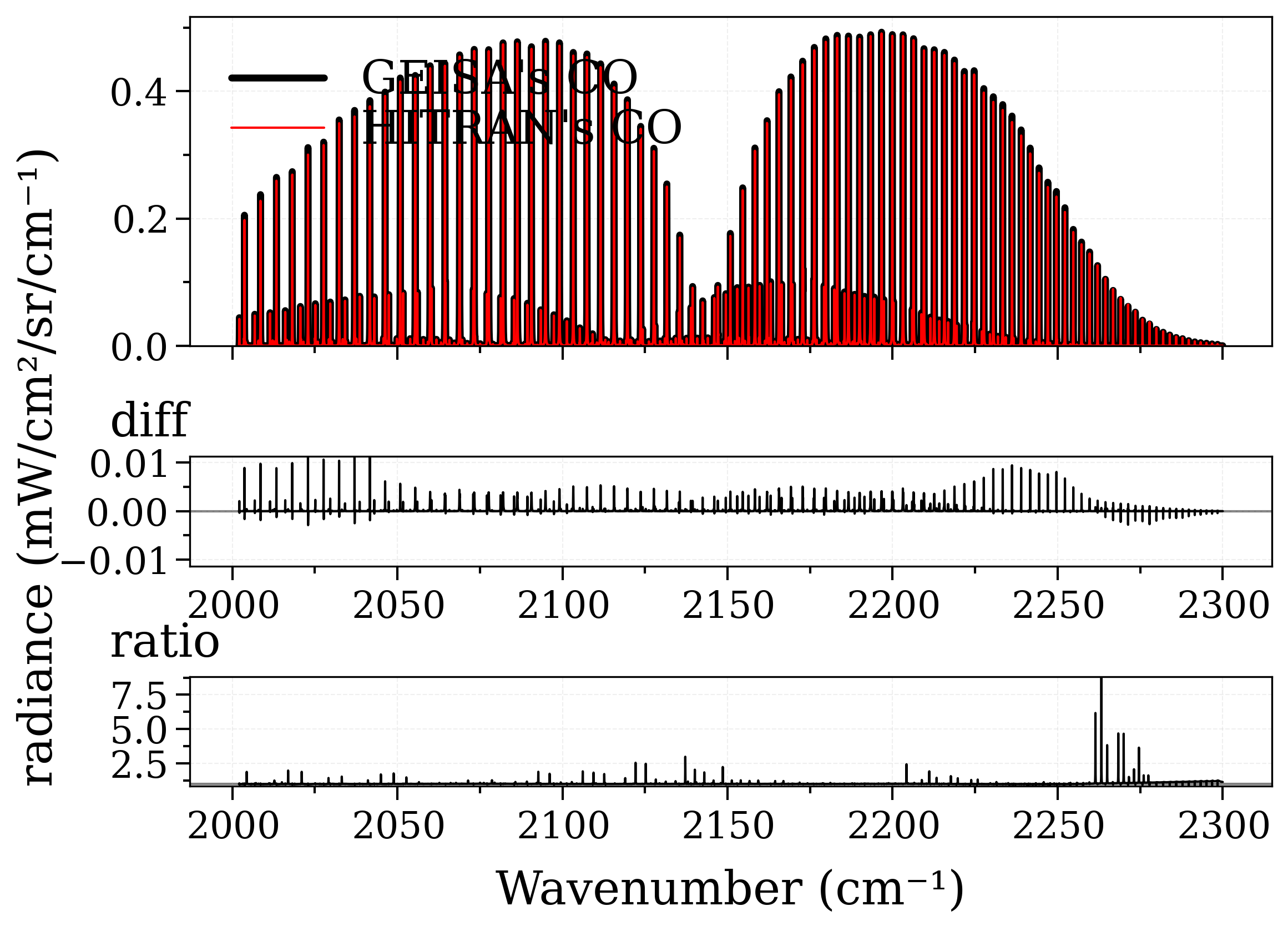
Auto-download and calculate CO spectrum from the GEISA database, and the HITRAN database.
Output should be similar, but not exactly! By default these two databases provide different broadening coefficients. However, the Einstein coefficients & linestrengths should be approximately the same, therefore the integrals under the lines should be similar.
You can see it by running the code below.
For your interest, GEISA and HITRAN lines can be downloaded and accessed separately using
fetch_geisa() and fetch_hitran()
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1,2,3,4,5,6
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2300.0000 cm-1
wavenum_min 2002.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat geisa
dbpath /home/docs/.radisdb/geisa/CO-line_GEISA2020_asc_gs08_v1.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 29802
----------------------------------------
0.06s - Spectrum calculated
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/db/molparam.py:252: FutureWarning:
The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
/home/docs/checkouts/readthedocs.org/user_builds/radis/checkouts/develop/radis/misc/warning.py:427: HighTemperatureWarning:
HITRAN is valid for low temperatures (typically < 700 K). For higher temperatures you may need HITEMP or CDSD. See the 'databank=' parameter
Calculating Equilibrium Spectrum
Physical Conditions
----------------------------------------
Tgas 1000 K
Trot 1000 K
Tvib 1000 K
isotope 1,2,3,4,5,6
mole_fraction 0.1
molecule CO
overpopulation None
path_length 1 cm
pressure 1.01325 bar
rot_distribution boltzmann
self_absorption True
state X
vib_distribution boltzmann
wavenum_max 2300.0000 cm-1
wavenum_min 2002.0000 cm-1
Computation Parameters
----------------------------------------
Tref 296 K
add_at_used numpy
broadening_method voigt
cutoff 1e-27 cm-1/(#.cm-2)
dbformat hitran
dbpath /home/docs/.radisdb/hitran/CO.hdf5
folding_thresh 1e-06
include_neighbouring_lines True
memory_mapping_engine auto
neighbour_lines 0 cm-1
optimization simple
parfuncfmt hapi
parsum_mode full summation
pseudo_continuum_threshold 0
sparse_ldm True
truncation 50 cm-1
waveunit cm-1
wstep 0.01 cm-1
zero_padding 29802
----------------------------------------
0.06s - Spectrum calculated
(<Figure size 640x480 with 3 Axes>, [<Axes: >, <Axes: >, <Axes: xlabel='Wavenumber (cm⁻¹)'>])
import astropy.units as u
from radis import calc_spectrum, plot_diff
conditions = {
"wmin": 2002 / u.cm,
"wmax": 2300 / u.cm,
"molecule": "CO",
"pressure": 1.01325, # bar
"Tgas": 1000, # K
"mole_fraction": 0.1,
"path_length": 1, # cm
"verbose": True,
}
s_geisa = calc_spectrum(**conditions, databank="geisa", name="GEISA's CO")
s_hitran = calc_spectrum(
**conditions,
databank="hitran",
name="HITRAN's CO",
)
"""
In :py:func:`~radis.io.geisa.fetch_geisa`, you can choose to additionally plot the
absolute difference (method='diff') by default, or the ratio (method='ratio'), or both.
"""
plot_diff(s_geisa, s_hitran, method=["diff", "ratio"])
Total running time of the script: (0 minutes 1.596 seconds)



